Abstract
The equilibrium geometries and chemical reactivities of the novel coumarine derivative, 3-[1-(3-hydroxypropylamino)ethylidene]chroman-2,4-dione, in water and benzene were investigated. The Fukui parameters, calculated by the Natural and Atoms in Molecules charges, were determined for all atoms in both phases. The most potent sites for the electrophilic, nucleophilic, and radical attack are discussed. Molecular docking analysis was carried out to identify the potency of inhibition of the title molecule against human cartilage proteins. The inhibition activity was obtained for ten conformations of ligand inside protein. This study proved that the Fukui indices can be used as the reactivity descriptors for the novel substances with inhibitory activity.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coumarine (2H-1-benzopyran-2-one) is a naturally occurring substance found in essential oils [1], sweet clover, and woodruff of cassia [2]. The significant amounts can enter human body through dietary exposure, because these compounds are found in fruits [3], vegetables [4, 5], seeds [6], trees [7], coffee, and vine [8]. The function of coumarins in plants is still a matter of dispute, although it is suggested that they can be involved in growth regulation, photosynthesis, and control of respiration [9], or behave as fungistats and bacteriostats [10].
The structure of coumarine (Fig. 1) contains fused pyrone and benzene rings along with the carboxyl group on the pyrone ring. The large group of coumarine derivatives can be divided into four sections: simple coumarins, pyranocoumarins, furanocoumarins, and the pyrone-substituted coumarins [8]. There are numerous techniques for the synthesis of the novel coumarine derivatives, that have been found to be applicable in medicine and pharmacy [11]. Coumarine derivatives can be used as antibreast cancer substances [11, 12], anti-HIV agents [6, 7], neuroprotectors, antivirals, anticonvulsants, antioxidants [9, 13, 14], antifungals [15], antibacterials [13], and antimutagens [16]. The most important use of coumarine derivatives in clinical practice is their application as anticoagulant agents [17, 18]. The application of coumarine and its derivatives also includes fluorescent probes [19,20,21], dyes [22, 23], and food additives [24]. There are also negative effects reported for the consumption of these compounds, including toxic and carcinogenic behavior [25, 26].
Various computational methods have been employed for the investigation of the activity of coumarine and its derivatives. Quantitative structure–activity relationship (QSAR) method is very popular for the elucidation of the structure–activity relationship towards different radicals [27,28,29]. QSAR and density functional theory (DFT) was used on the series of 4-hydroxychromene-2-one derivatives to evaluate their antiradical activity [30]. Molecular docking studies have proved as very important for the interactions of coumarine derivatives with biologically important proteins [12, 31,32,33,34].
Two cartilage proteins, fibrillin and chondroadherin were chosen for molecular docking study because of their biological importance. Fibrillin appears in cartilage as early as 20th week of fetal gestation. The loose bundles of microfibrils are formed until early adolescence, after which broad banded fibers can be found accumulated pericellulary within cartilage. These fibers can be extracted using dissociative conditions and it has been postulated that they are laterally packed and crosslinked microfibrils. The growth-regulating function of Fib-1 may reside persistently within the perichondrium. The accumulation of special laterally crosslinked Fib-1 microfibrils around chondrocytes during late adolescence led to the conclusion that growth-regulating activities may also be performed by Fib-1 at these sites [35]. The other docked protein, chondroadherin is a cartilage matrix protein. It is thought that this protein mediates adhesion of isolated chondrocytes. Chondroadherin is consisting of 11 leucine-rich repeats flanked by cysteine-rich regions. The RNA messenger for chondroadherin messenger RNA is present in chondrocytes at all ages. Further research is needed for determination which of its structural features responsible for cell binding and matrix-binding properties. The interactions of this protein with matrix components in vivo also require further investigation. The role in mediating cell–matrix interactions makes chondroadherin a potentially important regulator of chondrocyte organization within cartilage. Therefore, it can be speculated that abundance or deficiency of this protein cartilage function [36].
In this contribution, the novel coumarine derivate 3-[1-(3-hydroxypropylamino)ethylidene]chroman-2,4-dione (1), is investigated for the reactivity toward cartilages proteins by the means of the Natural Bond Orbital (NBO), Quantum Theory of Atoms in Molecules (QTAIM), and Molecular Docking analysis. The reactivity indices for specific atoms are also discussed.
Results and discussion
The optimized geometries in water and benzene of 1 are given in Fig. 2 (coordinates given in Tables S1 and S2; supplementary material). From the structural point of view there are no significant differences in these two geometries in solvents. The coumarine part and double bond C=N are planar. This is expected because the starting structure is very rigid. The optimized structure in water is more stable by 10.25 kJ mol−1 when energies with zero point corrections are compared. The enthalpy and Gibbs free energy in water is lower for 10.79 and 5.40 kJ mol−1, respectively. The energy difference is attributed to the interaction of polar bonds (N–H and O–H) with the polarizable continuum of solvent.
The HOMO–LUMO gap [(ΔE), Fig. 3] determines chemical reactivity and molecular stability. ΔE is directly related to the easiness of electron excitation of investigated molecule. Data from Fig. 3 for ΔE suggest that 1 has higher chemical activity in benzene than in water, since it has the slightly lower value of the energy gap (4.56 eV compared to 4.63 eV). From the visualized orbitals, it is possible to conclude that the orbitals are localized on the polar bonds and electronegative atoms in molecule. This leads to the higher stabilization of the orbitals as reflected in their energy. When structures in water and benzene are compared, the HOMO orbital is more stabilized in polar solvent than LUMO orbital, 0.15 and 0.08 eV, respectively, therefore the energy difference reflects the variation in electron density for specific orbitals with solvent polarity.
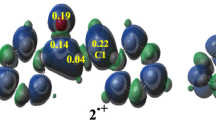
The overall reactivity of the molecule can be complemented with the reactivity of the specific sites for different types of reactions. To evaluate the condensed Fukui functions, single point calculations have been performed for the systems N (neutral molecule), N − 1 (radical cation), and N + 1 (radical anion) electrons, over the optimized neutral geometries using the unrestricted B3LYP formalism for radical species, and restricted B3LYP for neutral molecule in both solvents. Therefore, charge of 0, +1, and −1 is distributed over all of the atoms based on their electronegativity and position in molecule. The condensed Fukui functions were estimated from the NBO and AIM charges employing the Eq. (5). These two methods are often used for the calculation of Fukui functions (f) [37]. Table 1 gives the most probable sites for the electrophilic (f −), nucleophilic (f +), and radical attack (f 0) in water and benzene. The complete list of values for CFF in two solvents is given in the supplementary material. The numeration follows the positions from Fig. 2.
The highest value of f should mark the most probable site for the selected type of attack. Calculated values for the Fukui functions f, when two different methods are used, show notable differences in value. This is expected because the charges are obtained using different approaches. Generally speaking higher values for Natural charges are obtained, while values for AIM charges are lower. This is unexpected result, because the values of AIM charges are usually overestimated [38]. The authors have to note that the calculated values are dependent on the functional and basis set used, especially when NBO charges are concerned [39]. What is important to observe is that the atoms for electrophilic, nucleophilic, and radical attack are the same, with few exceptions, for both methods. The relative order shows resemblance but cannot be taken as absolute. The following discussion is based on the comparison of the four atoms with highest values of f in each column (also made bold in Table 1). The C4, C1′, O4, and C7 are the most reactive atoms for electrophilic attack in both solvents, with the values that are very similar. The NBO predicts that the C4 atom is the most reactive site in water while AIM predicts O4. On the other hand C1′ and O4 are the most reactive centers in benzene by NBO and AIM charges, respectively.
As for the nucleophilic attack, on the basis of the value of the NBO charges, three positions favored are C3, O2, and N1 in both solvents, while values of CFF calculated by AIM approach predict C3, O2, and C4 and C3, O2, and N1 in water and benzene, respectively. According to the NBO values C3 is the most reactive atom, while AIM predict O2 as the most reactive position in both solvents. It should be noted that the value of Fukui function on C3 (0.314 and 0.254) is significantly higher than for the other atoms thus proving the high reactivity in both solvents. The Fukui parameters calculated by NBO for the free-radical attack are within 0.5 units which proves the competitiveness of various positions, although C3 can be considered as the most reactive one. On the other hand, the values obtained using AIM charges predict that the most reactive atoms for the reaction with free-radical are carbonyl groups. The O2 and O4 are the most reactive in water while O2 is in benzene. It is obvious that the NBO determines the most reactive positions for BDE and/or SPLET mechanisms, while AIM predicts most reactive positions for RAF mechanism.
When different solvents are investigated and CFFs compared, the values of f depend on the polarity through the separation of charges. In the most cases the values of Fukui functions are lower in benzene. The reactive sites are the same as mentioned earlier, which shows that these predictions are not solvent-dependent. There are just few examples when the four most probable positions are not the same for two solvents—for example, C3 in water is excluded in benzene, while N1 is the group of possible positions for radical attack in benzene. The values of f functions between C3 and N1 for radical attack are not significantly different; therefore, it is ambiguous to distinguish which one of them is more probable position for attack. The influence of solvent polarity is more prominent for the nucleophilic attack then for electrophilic, which is in good accordance with the findings from the ref. [40].
As it can be observed in Tables 2 and S3–S4 (Supplementary material), some of the CFF have negative values. In the paper by Bultinck and coworkers [41] it is discussed that the negative values of CFF might indicate the short inter-atomic distances between atoms inside molecule. The values of CFF depend on the distance between atoms, as shown by Senthilkumar and Kolandaivel [38], and they tend towards negative as distance decreases.
To evaluate the inhibitory nature of 1 against human cartilage proteins (fibrillin and chondroadherin) molecular docking was performed. Protein–ligand binding energy and identification of the potential ligand binding sites were determined from this study as well. The ligand conformation which showed the lowest binding energy (best position) was determined based on the ligand docking results. The position and orientation of ligand inside protein receptor and the interactions with amino acids which bound to the ligand were analyzed and visualized with Discovery Studio 4.0 and AutoDockTools.
In Tables 2 and S5 (Supplementary material) values of estimated free energy of binding and inhibition constant (K i ) for the investigated ligand in ten different conformations are given. Lower value of K i indicates better inhibition.
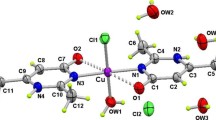
The lowest values of ΔG bind and K i are found for conformation 1 (Table 2; Table S5). Analyzing position of active amino acids, it can be concluded that ligand binds at the catalytic site of substrates by weak non-covalent interactions. The most prominent are H-bonds, alkyl–π, π–π and π–amide interactions. The atoms that interact with amino acids are O2, O4, and N1 as most potent sites for reaction proved by the Fukui indices. Glycine and aspartic acid in positions 15 and 110 in primary structures of chain A have predominant role as active sites of fibrillin and chondroadherin proteins for inhibition action, regardless of the conformation of investigated ligand (Fig. 4). GLY15 forms two H-bonds (2.07 and 2.32 Å length) with O–H and amino group of the ligand, while ASN14 forms H-bond of 2.90 Å length with C=O group of the ligand (Fig. 4). On other hand, ASP110 and HYS108 form hydrogen bonds of 2.12 and 2.30 Å length with O–H and C=O groups of ligand. LYS134 forms weak alkyl–π interaction with benzene ring of the ligand (Fig. 4). This analysis is good accordance with the previous discussion, proving that the Fukui parameters can be used for the prediction of the active sites for biologically active molecules.
Conclusion
In present work the reactivity parameters for molecule 3-[1-(3-hydroxypropylamino)ethylidene]chroman-2,4-dione (1) were calculated at the B3LYP-D3BJ/6–311+G(d,p) level of theory in the aqueous (polar solvent) and benzene (non-polar solvent) phases. The HOMO–LUMO gap (ΔE) suggested that 1 has higher chemical activity in benzene than in water, because it has slightly lower value in non-polar solvent.
The condensed Fukui indices were calculated for all atoms in molecule 1 by the means of Natural and Atoms in Molecules charges. Based on these parameters, the most probable reactions sites for nucleophilic, electrophilic, and radical attack were determined. The C4, C7, O4, and C1′ atoms are favorable sites for electrophilic attack, while for nucleophilic attack the C3, O2, and N1 atoms are suitable. For free-radical attack, the C3, O2, and N1 atoms are possible positions for reaction.
The title ligand, according to the results of the molecular docking study, forms a stable complex with human cartilage proteins as evident from the binding energy (ΔG bind in kJ mol−1). The most important interactions are H-bonds, alkyl–π, π–π and π–amide. Atoms forming bonds are predicted as the most reactive sites by the Fukui indices. These preliminary results suggest that the investigated ligand 1 might exhibit inhibitory activity against fibrillin and chondroadherin proteins.
Methods
Chemical reactivity of molecule
The molecular structure and the electronic parameters, such as the energies of highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) and the HOMO–LUMO gap (∆E = E LUMO − E HOMO) can be obtained through theoretical calculations [42,43,44,45,46,47,48].
Fukui functions
Fukui function, proposed by Parr and Yang in 1984 [48,49,50], describes the differential change of the electron density induced by the change in total number of electrons. It is mathematically defined by the following equation:
In the previous equation µ and N are the chemical potential and the number of electrons in the system, υ(r) is the external potential. The Fukui function is often calculated approximatively as follows:
The evaluation of these equations can be very complicated due to the electron density term [47]. In Eq. (2), the frozen-orbital approximation is assumed. ρ N (r), ρ N−1(r), and ρ N+1(r) represent separately the electron densities of the system with N (neutral molecule), N − 1 (radical cation), and N + 1 (radical anion) electrons. The values of ρ HOMO(r) and ρ LUMO(r) indicate the electron densities of the HOMO and LUMO, respectively.
If Eq. (2) is integrated for individual atoms in a molecule, one obtains the so-called condensed Fukui function (CFF) [51], which provide a more convenient way to predict the reaction site in a molecule. The definition of condensed Fukui function for an atom, noted as A, can be written as:
where \(p_{N}^{\text{A}}\) is the electron population number of atom A. Since atomic charge is defined as q A = Z A − p A, where Z is the charge of atomic nucleus, f − and f + can be expressed as the difference of atomic charges in two states (note that the two Z terms are canceled). By analogous treatment, one can easily formulate the CFFs for an atom A in Eq. (5):
where \(q_{N}^{\text{A}}\), \(q_{N - 1}^{\text{A}}\), and \(q_{N + 1}^{\text{A}}\) are the charges at atom A of the neutral, anionic, and cationic species, respectively [51].
The three formulas in Eq. (5) are exploited in this work to describe the electrophilic, nucleophilic and free-radical attack. In general, the larger the value of the condensed Fukui function at a reaction site, the greater the reactivity of that corresponding site towards reactive species. There are various methods for the calculation of the atomic charges, many of which were employed for the Fukui indices [51,52,53]. In this paper the atoms in molecules (AIM) and natural charges were used for calculations of the CFF.
Computational methods
The geometries of neutral molecules (Fig. 2) were optimized by the density functional theory (DFT) using global hybrid generalized gradient approximation (GGA) functional B3LYP [54, 55] with empirical dispersion corrections D3BJ (with Becke and Johnson damping) [54,55,56,57,58,59] in combination with the 6–311+G(d,p) basis set. Many authors employed the B3LYP-D3BJ functional successfully for description of similar systems [57,58,59]. B3LYP-D3BJ was selected as a widely applicable method that proved to describe inter-atomic interactions, at short and medium distances (≤5 Å), more accurately and reliably than traditional DFT methods. Hybrid GGA B3LYP-D3BJ includes an empirical correction term proposed by Grimme [57]. The geometry optimization was performed in Gaussian 09 program package [60]. The nature of the stationary points was determined by performing frequency analysis: the equilibrium geometries had no imaginary vibrations. The solvent effect was taken into account in geometry optimization and energy calculation using the SMD implicit solvation model [61]. The applied SMD continuum solvation model is based on interaction between the charge density of solute molecule with dielectric medium. The geometry was optimized in water (polar solvent) and benzene (non-polar solvent) to investigate the effect of solvent polarity on reactivity parameters. The NBO analysis was performed in the NBO 5.9 software [62, 63]. Quantum theory of atoms in molecules (QTAIM) calculations was done based on the wfn files produced by the Gaussian and analyzed in the Multiwfn Package [64] for the atomic charges. The analysis of atomic basins was performed at the high quality grid with spacing of 0.06 B. The molecular docking simulation was carried out using the AutoDock 4.0 software [65]. The three-dimensional crystal structures of human fibrillin and chondroadherin were obtained from the Protein Data Bank (PDB IDs: 2W86 and 5LFN) [66, 67]. The Discovery Studio 4.0 [68] was used for preparation of protein for docking by removing the co-crystallized ligand, water molecules and co-factors. To calculate Kollman charges and to add polar hydrogen the AutoDockTools (ADT) graphical user interface was used. Title molecule 1 was prepared for docking by minimizing its energy at B3LYP-D3BJ/6–311+G(d,p) level of theory. The flexibility of the ligands is considered, while the protein or biomolecules remained as rigid structure in the ADT. All bonds of 1 were set to be rotatable. The Geistenger method for calculation of partial charges was employed. All calculations for protein—ligand flexible docking were done using the Lamarckian genetic algorithm (LGA) method. The grid boxes with dimensions 17.442 × 28.297 × 58.056 of fibrillin and 7.214 × 59.888 × 95.047 of chondroadherin were used to cover the protein binding site and accommodate ligand to move freely. Inhibition potency of title molecule was investigated and discussed.
References
Choi J, Lee K-T, Ka H, Jung WT, Jung HJ, Park HJ (2001) Arch Pharm Res 24:418
Cohen AJ (1979) Food Cosmet Toxicol 17:277
Mead JA, Smith JN, Williams RT (1958) Biochem J 68:67
Rosselli S, Maggio AM, Faraone N, Spadaro V, Morris-Natschke SL, Bastow KF, Lee KH, Bruno M (2009) Nat Prod Commun 4:1701
Rahman A, Shabbir M, Ziauddin-Sultani S, Jabbar A, Choudhary MI (1997) Phytochemistry 44:683
Spino C, Dodier M, Sotheeswaran S (1998) Bioorg Med Chem Lett 8:3475
Fuller RW, Bokesch HR, Gustafson KR, McKee TC, Cardellina JH II, McMahon JB, Cragg GM, Soejarto DD, Boyd MR (1994) Bioorg Med Chem Lett 4:1961
Venugopala KN, Rashmi V, Odhav B (2013) Biomed Res Int 2013:963248
Hatano T, Yasuhara T, Fukuda T, Noro T, Okuda T (1989) Chem Pharm Bull 37:3005
Bogdał D (1998) J Chem Res 468
Musa M, Cooperwood J, Khan MO (2008) Curr Med Chem 15:2664
Pingaew R, Saekee A, Mandi P, Nantasenamat C, Prachayasittikul S, Ruchirawat S, Prachayasittikul V (2014) Eur J Med Chem 85:65
Kayser O, Kolodziej H (1997) Planta Med 63:508
Fylaktakidou K, Hadjipavlou-Litina D, Litinas K, Nicolaides D (2004) Curr Pharm Des 10:3813
Hoult JRS, Payá M (1996) Gen Pharmacol Vasc Syst 27:713
Khode S, Maddi V, Aragade P, Palkar M, Ronad PK, Mamledesai S, Thippeswamy AH, Satyanarayana D (2009) Eur J Med Chem 44:1682
Lowenthal J, Birnbaum H (1969) Science 164:181
Hildebrandt EF, Suttie JW (1982) Biochemistry 21:2406
Soumya TV, Thasnim P, Bahulayan D (2014) Tetrahedron Lett 55:4643
Wagner BD (2009) Molecules 14:210
Ray D, Bharadwaj PK (2008) Inorg Chem 47:2252
Hara K, Kurashige M, Dan-oh Y, Chiaki K, Shinpo A, Suga S, Sayamaa K, Arakawa H (2003) New J Chem 27:783
Hara K, Sayama K, Ohga Y, Shinpo A, Suga S, Arakawa H (2001) Chem Commun 115:569
Akman F (2016) Can J Phys 94:583
Abraham K, Wöhrlin F, Lindtner O, Heinemeyer G, Lampen A (2010) Mol Nutr Food Res 54:228
Egan D, O’Kennedy R, Moran E, Cox D, Prosser E, Thornes RD (1990) Drug Metab Rev 22:503
Zhang H-Y, Wang L-F (2004) J Mol Struct Theochem 673:199
Lin HC, Tsai SH, Chen CS, Chang YC, Lee CM, Lai ZY, Lin CM (2008) Biochem Pharmacol 75:1416
Beillerot A, Domínguez J-CR, Kirsch G, Bagrel D (2008) Bioorg Med Chem Lett 18:1102
Mladenović M, Mihailović M, Bogojević D, Matić S, Nićiforović N, Mihailović V, Vuković N, Sukdolak S, Solujić S (2011) Int J Mol Sci 12:2822
Liu XH, Liu HF, Chen J, Yang Y, Song BA, Bai LS, Liu JX, Zhu HL, Qi XB (2010) Bioorg Med Chem Lett 20:5705
Stefani HA, Gueogjan K, Manarin F, Farsky SH, Zukerman-Schpector J, Caracelli I, Pizano Rodrigues SR, Muscará MN, Teixeira SA, Santin JR, Machado ID, Bolonheis SM, Curi R, Vinolo MA (2012) Eur J Med Chem 58:117
Delogu G, Picciau C, Ferino G, Quezada E, Podda G, Uriarte E, Viña D (2011) Eur J Med Chem 46:1147
Huang XY, Shan ZJ, Zhai HL, Su L, Zhang XY (2011) Chem Biol Drug Des 78:651
Keene DR, Jordan CD, Reinhardt DP, Ridgway CC, Ono RN, Corson GM, Fairhurst M, Sussman MD, Memoli VA, Sakai LY (1997) J Histochem Cytochem 45:1069
Grover J, Chen X-N, Korenberg JR, Roughley PJ (1997) Genomics 45:379
Parr RG, Pearson RG (1983) J Am Chem Soc 105:7512
Senthilkumar L, Kolandaivel P (2005) Mol Phys 103:547
Proft FD, Martin JML, Geerlings P (1996) Chem Phys Lett 256:400
Kar R, Pal S (2009) Int J Quantum Chem 110:1642
Bultinck P, Carbó-Dorca R, Langenaeker W (2003) J Chem Phys 118:4349
Parr RG (1980) Density functional theory of atoms and molecules. In: Fukui K, Pullman B (eds) Horizons of quantum chemistry. Springer, Dordrecht
Gázquez JL (2008) J Mex Chem Soc 52:3
Geerlings P, Proft FD, Langenaeker W (2003) Chem Rev 103:1793
Zevatskii YE, Samoilov DV (2007) Russ Org Chem 43:483
Parr RG, Donnelly RA, Levy M, Palke WE (1978) J Chem Phys 68:3801
Parr RG, Szentpály LV, Liu S (1999) J Am Chem Soc 121:1922
Parr RG, Yang W (1984) J Am Chem Soc 106:4049
Liu S (2009) Acta Phys Chim Sin 25:590
Yang W, Mortier WJ (1986) J Am Chem Soc 108:5708
Tiznado W, Chamorro E, Contreras R, Fuentealba P (2005) J Phys Chem A 109:3220
Becke AD (1988) Phys Rev A 38:3098
Becke AD (1993) J Chem Phys 98:5648
Becke AD, Johnson ER (2005) J Chem Phys 123:154101
Johnson ER, Becke AD (2006) Chem Phys Lett 432:600
Grimme S, Antony J, Ehrlich S, Krieg H (2010) J Chem Phys 132:154104
Grimme S, Ehrlich S, Goerigk L (2011) J Comput Chem 32:1456
Sardo M, Siegel R, Santos SM, Rocha J, Gomes JR, Mafra L (2012) J Phys Chem A 116:6711
Ivanov P (2016) J Mol Struct 1107:31
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukud R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JJA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brother E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi MR, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts RE, Stratmann O, Yazyev AJ, Austin R, Cammi C, Pomelli JW, Ochterski R, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision C.01. Gaussian Inc, Wallingford
Marenich AV, Cramer CJ, Truhlar DG (2009) J Phys Chem B 113:6378
Carpenter JE, Weinhold F (1988) J Mol Struct Theochem 169:41
Reed AE, Curtiss LA, Weinhold F (1988) Chem Rev 88:899
Lu T, Chen F (2012) J Comput Chem 33:580
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) J Comput Chem 30:2785
Jensen SA, Iqbal S, Lowe ED, Redfield C, Handford PA (2009) Structure 17:759
Rämisch S, Pramhed A, Tillgren V, Aspberg A, Logan DT (2017) Acta Crystallogr D Struct Biol 73:53
BIOVIA Discovery Studio v4.0. Accelrys Software Inc, San Diego, 2005-2013
Acknowledgements
This work was supported by the Ministry of Science of the Republic of Serbia (Projects nos. OI172015, OI172016, and OI174028).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Milenković, D., Avdović, E.H., Dimić, D. et al. Reactivity of the coumarine derivative towards cartilage proteins: combined NBO, QTAIM, and molecular docking study. Monatsh Chem 149, 159–166 (2018). https://doi.org/10.1007/s00706-017-2051-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00706-017-2051-4