Abstract
A newly identified lytic Cronobacter phage, GW1, was isolated from the Pearl River of Guangzhou, China. GW1 had a double-stranded DNA genome of 39,695 nucleotides with an average GC content of 53.18 %. Among the 49 open reading frames (ORFs) identified, genes for rRNA, tRNA, antibiotic resistance, and virulence factors were not found in the phage genome. The morphology, genomic features, and phylogenetic position of GW1 revealed that it represents a new species in the genus T7virus. This novel lytic Cronobacter phage may provide an alternative for phage therapy and biocontrol against Cronobacter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cronobacter has been isolated from various foods, environments, and clinical samples, and in particular from reconstituted infant milk formula [2, 18]. Currently, the genus Cronobacter comprises seven species: Cronobacter sakazakii, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter universalis, Cronobacter muytjensii, Cronobacter dublinensis, and Cronobacter condimenti [8]. These species are opportunistic foodborne human pathogens that can cause infections in all age groups [11]. Clinical symptoms of infection in infants include meningitis, bacteremia, and necrotizing enterocolitis, with mortality rates of up to 40%-80% [5].
Bacteriophages, viruses that infect host bacteria, are extremely abundant in many environments [16]. The use of bacteriophages as novel biological control agents to decrease food pathogen outbreaks and enhance food safety has been reported in recent years [6]. The high genetic diversity of Cronobacter is a challenge to the use of phages for controlling microbial contamination in food and the food-processing environment. Isolation and characterization of more novel phages, especially lytic phages of Cronobacter, is necessary for phage research.
In this study, water samples were obtained from the Pearl River of Guangzhou, China. The Cronobacter turicensis cro1541A1-1 strain was used as the host for phage isolation. The isolation and enrichment procedures for the target phages were performed as described previously with some modifications [4]. One of the phages, named GW1, was isolated from a single plaque and was purified at least 10 times via plaque assay. The host range was determined using a standard plaque assay as reported previously [14].
Genomic DNA of phage GW1 was extracted using phenol and chloroform and sequenced using the Ion torrent S5 platform (Thermo Fisher Scientific, USA). The complete genome sequence was compared with other nucleotide sequences using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and PASC (https://www.ncbi.nlm.nih.gov/sutils/pasc/) at NCBI. Potential open reading frames (ORFs) were predicted using PHASTER (http://phaster.ca/). The putative functions of the proteins encoded by the ORFs were annotated using BLASTp against the Non-Redundant Protein Database of NCBI [3]. Putative rRNA and tRNA genes were analyzed using RNAmmer (http://www.cbs.dtu.dk/services/RNAmmer/) and tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/), respectively. All annotated GW1 genes were compared against the Antibiotic Resistance Genes Database (ARDB, https://card.mcmaster.ca/) and the virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/). A map representing the phage GW1 genome was generated using the CGView Server (http://stothard.afns.ualberta.ca/cgview_server/). A neighbor-joining tree was constructed using MEGA 7.0 [10].
Phage GW1 forms clear and round plaques with a diameter ranging from 4 to 6 mm after 8 h of incubation at 37°C, indicating that it is a lytic phage (Fig. S1). Electron microscopy revealed that GW1 belongs to the family Podoviridae. The head and short tail of the phage were found to be approximately 55 ± 3 nm and 16.6 ± 1 nm long, respectively (Fig. S2).
The possible host range of phage GW1 was tested using 15 Cronobacter strains covering five species. These strains included C. sakazakii (cro810A3, cro2810A3, and cro7), C. malonaticus (cro914W, cro138, and cro1064W), C. dublinensis (cro1046W, cro3314A1, and cro981W), C. muytjensii (cro1187A3, cro392B3, and cro1187W), and C. turicensis (cro1541A1-1, cro2864C1, and cro2991W) isolates (Table S1). The results showed that GW1 can grow on four strains belonging to C. turicensis (cro1541A1-1), C. muytjensii (cro1187W), and C. malonaticus (cro914W and cro138).
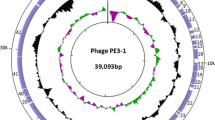
According to the International Committee on the Taxonomy of Viruses (ICTV), the main species demarcation criterion for defining new species of bacterial and archaeal viruses is currently set at a genome sequence identity of 95%. This indicates that two viruses belonging to the same species should differ from each other by less than 5% at the nucleotide level using several analytical tools such as BLASTn or PASC [1]. The 39,695-bp-long GW1 genome and was compared with those of other phages. In BLASTn, the data revealed that the genome of phage GW1 had the highest identity of 94% (91% coverage) to Citrobacter phage SH4 (KU687350.1), followed by 93% identity (92% coverage) to Cronobacter phage Dev2 (HG813241.1) [9]. The result in PASC showed that phage GW1 shared the highest identity of 86.87% with Cronobacter phage Dev2. Furthermore, the GW1 genome was found to have a GC content of 53.18%, which is higher than the 52.60% and 52.16% GC content in Citrobacter phage SH4 and Cronobacter phage Dev2, respectively. These results suggest that GW1 is a novel phage belonging to the genus T7virus.
Subsequently, 49 ORFs were identified in the genome, and they were all predicted to be transcribed from the same DNA strand. Based on their high sequence similarity to known proteins in GenBank, 27 ORFs were predicted to encode proteins (Table S2), that are mainly related to phage structure, host lysis, and DNA metabolism. Genes for rRNA, tRNA, antibiotic resistance, and virulence factors were not detected in the GW1 genome sequence.
As shown in Fig. 1, the predicted proteins related to host lysis mainly consisted of gp3, gp5, and gp28 [19]. Of note, gp3 and gp28 had sequence similarity to T7 holin and lysin, respectively. The holin and lysin proteins have been shown to play a significant role in lysing their host cells [20, 23]. Proteins (gp16, gp21, gp24, gp25, gp26, gp30, gp32, gp33, and gp36) encoded by the GW1 genome were predicted to be involved in DNA metabolism [7, 13, 15, 21, 22]. In addition, ten proteins and nine proteins were unique to GW1 when compared to SH4 and Dev2, respectively, and 60% of these unique proteins belonged to early-expressed genes (Fig. 1). The gp13 of GW1, which is predicted to be a protein kinase, is not present in SH4. This protein has been reported to play an important role in viral reproduction under specific suboptimal growth conditions [12]. The gp9 of phage GW1, annotated as S-adenosyl-L-methionine hydrolase (SAMase) since it shared 62% identity with Enterobacter phage T3 (KC960671.1) gp0.3 (SAMase), was also not observed in phages SH4 and Dev2. SAMase is responsible for overcoming the restriction-modification (R-M) system of the host [17].
Schematic representation of the genomic organization of phage GW1 compared to phages SH4 and Dev2. The genome is divided into an early-expressed region (green), a middle-expressed region (blue), and a late-expressed region (red) based on their homologs in members of the genus T7virus. Differences in shading indicate the percent amino acid sequence identity of the ORFs. Arrows indicate functional proteins, and rectangles represent hypothetical proteins
The available amino acid sequences of DNA polymerase and RNA polymerase from some closely related phages (more than 90% identity) including Citrobacter phage SH4, Cronobacter phage Dev2, and Citrobacter phage SH5 (KU687351.1), and those of other related phages such as Citrobacter phage SH3 (KU687349.1), Escherichia phage vB EcoP F (KY295894.1), Escherichia phage JSS1 (KX689784.2), Enterobacter phage E-4 (KP791807.1), Yersinia pestis phage phiA1122 (AY247822.1), Enterobacter phage T3 (KC960671.1), Enterobacter phage T7 (GU071091.1), Cronobacter phage Dev CD 23823 (LN878149.1), and Cronobacter sakazakii phage vB CskP GAP227 (KC107834.1) were downloaded from NCBI for phylogenetic analysis. The phylogenetic trees based on DNA polymerase and RNA polymerase are shown in Fig. 2. In the DNA polymerase tree, phage GW1 was grouped with SH4, Dev2, and SH5 into one large cluster; however, it was still distinct from these phages (Fig. 2A). More importantly, in the RNA-polymerase-based tree (Fig. 2B), phage GW1 was located in an outgroup of six phages belonged to different species of the genus T7virus. This indicated a clear distant phylogenetic relationship to these identified phages.
In conclusion, combined with the less than 95% genome sequence identity, the presence of ORFs that are lacking in the similar phages SH4 and Dev2 and its distant phylogenetic relationship to known phages indicate that phage GW1 is a member of a new species in the genus T7virus. Characterization of the properties of GW1 is an important prerequisite for its further application in the food industry.
Nucleotide sequence accession number
The GenBank accession number of the whole genome sequence of Cronobacter phage GW1 reported in this article is MH491167.
References
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses. https://doi.org/10.3390/v9040070
Alsonosi A, Hariri S, Kajsik M, Orieskova M, Hanulik V, Roderova M, Petrzelova J, Kollarova H, Drahovska H, Forsythe S, Holy O (2015) The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur J Clin Microbiol Infect Dis 34(10):1979–1988. https://doi.org/10.1007/s10096-015-2440-8
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Capra ML, Del LQA, Ackermann HW, Moineau S, Reinheimer JA (2006) Characterization of a new virulent phage (MLC-A) of Lactobacillus paracasei. J Dairy Sci 89(7):2414–2423. https://doi.org/10.3168/jds.S0022-0302(06)72314-1
Friedemann M (2009) Epidemiology of invasive neonatal Cronobacter (Enterobacter sakazakii) infections. Eur J Clin Microbiol Infect Dis 28(11):1297–1304. https://doi.org/10.1007/s10096-009-0779-4
Garcia P, Martinez B, Obeso JM, Rodriguez A (2008) Bacteriophages and their application in food safety. Lett Appl Microbiol 47(6):479–485. https://doi.org/10.1111/j.1472-765X.2008.02458.x
Ikeda RA, Richardson CC (1986) Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc Natl Acad Sci USA 83(11):3614–3618
Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ (2012) Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol 50(9):3031–3039. https://doi.org/10.1128/JCM.00905-12
Kajsík M, Oslanecová L, Szemes T, Hýblová M, Bilková A, Drahovská H, Turňa J (2014) Characterization and genome sequence of Dev2, a new T7-like bacteriophage infecting Cronobacter turicensis. Arch Virol 159(11):3013–3019
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Liu H, Cui JH, Cui ZG, Hu GC, Yang YL, Li J, Shi YW (2013) Cronobacter carriage in neonate and adult intestinal tracts. Biomed Environ Sci BES 26(10):861–864. https://doi.org/10.3967/bes2013.011
Michalewicz J, Nicholson AW (1992) Molecular cloning and expression of the bacteriophage T7 0.7(protein kinase) gene. Virology 186(2):452–462
Nakai H, Richardson CC (1986) Interactions of the DNA polymerase and gene 4 protein of bacteriophage T7. Protein-protein and protein-DNA interactions involved in RNA-primed DNA synthesis. J Biol Chem 261(32):15208–15216
O’Flaherty S, Ross RP, Flynn J, Meaney WJ, Fitzgerald GF, Coffey A (2005) Isolation and characterization of two anti-staphylococcal bacteriophages specific for pathogenic Staphylococcus aureus associated with bovine infections. Lett Appl Microbiol 41(6):482–486. https://doi.org/10.1111/j.1472-765X.2005.01781.x
Owen-Hughes TA, Pavitt GD, Santos DS, Sidebotham JM, Hulton CS, Hinton JC, Higgins CF (1992) The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71(2):255–265
Parikka KJ, Le Romancer M, Wauters N, Jacquet S (2017) Deciphering the virus-to-prokaryote ratio (VPR): insights into virus-host relationships in a variety of ecosystems. Biol Rev Camb Philos Soc 92(2):1081–1100. https://doi.org/10.1111/brv.12271
Studier FW, Movva NR (1976) SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J Virol 19(1):136–145
Ueda S (2017) Occurrence of Cronobacter spp. in dried foods, fresh vegetables and soil. Biocontrol Sci 22(1):55–59. https://doi.org/10.4265/bio.22.55
Wang Q, Zeng X, Yang Q, Yang C (2018) Identification of a bacteriophage from an environmental multidrug-resistant E. coli isolate and its function in horizontal transfer of ARGs. Sci Total Env 639:617–623. https://doi.org/10.1016/j.scitotenv.2018.05.213
Young I, Wang I, Roof WD (2000) Phages will out: strategies of host cell lysis. Trends Microbiol 8(3):120–128
Yuan L, Cui Z, Wang Y, Guo X, Zhao Y (2014) Complete genome sequence of virulent bacteriophage SHOU24, which infects foodborne pathogenic Vibrio parahaemolyticus. Arch Virol 159(11):3089–3093. https://doi.org/10.1007/s00705-014-2160-x
Zhang X, Studier FW (2004) Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J Mol Biol 340(4):707–730. https://doi.org/10.1016/j.jmb.2004.05.006
Ziedaite G, Daugelavicius R, Bamford JK, Bamford DH (2005) The Holin protein of bacteriophage PRD1 forms a pore for small-molecule and endolysin translocation. J Bacteriol 187(15):5397–5405. https://doi.org/10.1128/JB.187.15.5397-5405.2005
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31601571), the National Key R&D Program of China (2017YFC1601200), Pearl River S&T Nova Program of Guangzhou (201806010062), the Natural Science Foundation of Guangdong Province (2016A030310315), and GDAS’ Special Project of Science and Technology Development (2017GDASCX-0201). All authors read and approved the final manuscript. None of the authors reported a conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Chan-Shing Lin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zeng, H., He, W., Li, C. et al. Complete genome analysis of a novel phage GW1 lysing Cronobacter. Arch Virol 164, 625–628 (2019). https://doi.org/10.1007/s00705-018-4084-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4084-3