Abstract
The genome of a Cronobacter sakazakii M1 phage named PF-CE2 was characterized in this work, and a new species named "Cronobacter virus PF-CE2", in the genus Pseudotevenvirus of the subfamily Tevenvirinae of the family Myoviridae is proposed. The Gp190 gene of phage PF-CE2 is predicted to encode a bacteriophage-borne glycanase that is capable of degrading fucose-containing exopolysaccharides produced by C. sakazakii M1. Furthermore, we propose changing the taxonomic status of eight additional phages based on nucleotide sequence comparisons. This work provides a theoretical basis for subsequent heterologous expression of the phage PF-CE2 glycanase and provides an important reference for the preservation and sharing of these phages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Cronobacter sakazakii are facultative, anaerobic Gram-negative bacteria that are present in various foods and raw materials [1, 2]. In recent years, a new foodborne pathogen, C. sakazakii, has been commonly found in formula milk powder and is associated with several infectious diseases, including meningitis, necrotizing enterocolitis, and sepsis [3, 4]. Viruses are ubiquitous in nature, and bacteriophages, which are viruses of bacteria, are effective tools for killing pathogenic bacteria [5]. The genetic diversity of C. sakazakii poses a challenge for the use of phages to control microbial contamination in food-processing environments, and it is therefore necessary to isolate and identify new phages targeting C. sakazakii [6].
Fucose-containing exopolysaccharides (FcEPSs) are a promising source of fucosylated oligosaccharides and fucose. Cronobacter spp. typically have the capacity to produce fucose-rich FcEPSs [7]. Bacteriophage-borne glycanases extracted from phages are effective tools for degrading FcEPSs [8]. A previous study found that a phage isolated from sewage was capable of degrading FcEPS produced by C. sakazakii M1 [9]. In this study, a bacteriophage targeting C. sakazakii M1 was purified via more than 10 rounds of single-plaque isolation. The genome sequence and functional biological characteristics of phage PF-CE2 were determined and compared with those of homologous phages. In addition, a gene encoding a putative bacteriophage-borne glycanase was identified.
Phage isolation and purification were performed as described previously [9]. Briefly, prior to phage PF-CE2 DNA isolation, DNase I (10 μg/mL, Sigma-Aldrich) and RNase A (20 μg/mL, Sigma-Aldrich) were added to a purified suspension of PF-CE2 and incubated at 37 °C for 1 h to digest bacterial DNA and RNA. DNA isolation and purification of phage PF-CE2 were carried out using an E.Z.N.A® Viral DNA Kit (OMEGA). A sequencing library was generated using an Illumina TruSeq DNA Nano Sample Preparation Kit (Illumina). One microgram of DNA was sheared into 300- to 500-bp fragments using an M220 Focused-ultrasonicator (Covaris). After PCR amplification, specific bands were recovered by gel excision. A TBS-380 Micro-Fluorometer (Turner BioSystems) and PicoGreen® (Thermo Fisher Scientific) were used for quantitative analysis, and clusters were generated by bridging PCR amplification on a cBot 2 system (Illumina). The genome of phage PF-CE2 was sequenced using an Illumina HiSeq system with a 2 × 150-bp paired-end run. To ensure the accuracy and reliability of the sequencing results, quality control of the original data was performed as follows: (1) the adapter sequence was removed from reads, (2) reads containing non-AGCT at the 5' end were removed, (3) the ends of reads whose sequencing quality was less than Q20 were removed, (4) reads whose N proportion was more than 10% were removed, and (5) fragments less than 50 bp were discarded. Following quality control, clean data were obtained; detailed information is shown in Supplementary Table S1. The reads were assembled using ABySS (v2.0.2) assembly software, and GapCloser (v1.12) software was used to carry out gap filling and base correction.
The results of high-throughput sequencing showed that phage PF-CE2 was assembled at 88-fold coverage into a genome of 178,248 bp in length with a G + C content of 44.8% and that protein-encoding regions made up 95.87% of the genome. The genome sequence of phage PF-CE2 was compared with that of other phages using the standard nucleotide BLAST in the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Supplementary Table S2 shows the basic characteristics of eight selected phages, which are similar to phage PF-CE2 in length and G + C content, including the Citrobacter phages Margaery (unpublished) and Maroon [10], the Cronobacter phages vB CsaM Cronuts (unpublished), vB CsaM GAP161 [11], vB CsaM leB [1], vB CsaM leE [1], and vB CsaM leN [1], and the Enterobacter phage vB EkoM5VN (unpublished). tRNAscan-SE 2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/) was used to identify regions encoding possible tRNAs in the genomes of these phages, and rRNA was predicted using the RNAmmer 1.2 Server (http://www.cbs.dtu.dk/services/RNAmmer/) [12, 13]. None of them contained rRNA, tRNAMet and tRNAGly were found in the genomes of PF-CE2, Cronobacter phages vB CsaM Cronuts, and vB CsaM GAP161, and the others contained only tRNAGly. Comparisons to the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/) and Virulence Factor Database (http://www.mgc.ac.cn/VFs/blast/blast.html) did not detect antibiotic resistance genes or virulence factors in any of the genomes.
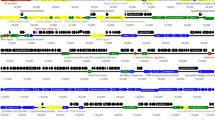
A total of 275 open reading frames (ORFs) were identified using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/), of which 123 were located on the plus strand and the remainder were on the minus strand (Supplementary Table S3). BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to search for sequences homologous to the 275 ORFs. A circular representation of the phage PF-CE2 genome was generated using the BLAST Ring Image Generator (BRIG) [14]. As shown in Fig. 1, the genome consisted of several clusters, encoding structural proteins, DNA replication and transcription proteins, nucleotide metabolism and biosynthesis proteins, lysis proteins, and DNA packaging proteins.
Schematic representation of the genomic organization of phage PF-CE2 compared to eight other selected phages. The outermost circle represents the predicted protein functions of coding sequences: structural proteins (blue), replication and transcription (orange), DNA nucleotide metabolism and biosynthesis (yellow), lysis proteins (purple), DNA packaging (black), others (fuchsia).
The above 275 ORFs were analyzed using the CAZy database (http://www.cazy.org/). The results showed that only the protein encoded by Gp190 was assigned to the GH family, which implies that Gp190 may have a function related to the degradation of exopolysaccharides. To validate this hypothesis, Gp190 was analyzed using PSI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) [15], and the results of the two iterations are shown in Supplementary Table S4. In the first iteration, 33 homologous glycoside hydrolase family protein sequences were found in the “Sequence with E-values WORSE than the threshold”. In the second iteration, 45 homologous sequences associated with glycoside hydrolase family proteins were shown in “Sequence with E-values BETTER than threshold”. In addition, HHPred (https://toolkit.tuebingen.mpg.de/tools/hhpred) was also used to analyze the potential function of Gp190. The results showed that the N-terminus of Gp190 was similar to the glycoside hydrolase catalytic center of 1WTH, with a probability of 100% [16], and that the C-terminus was similar to the glycoside hydrolase catalytic center of 3A1M, with a probability of 94.4% [17]. Bacteriophage-borne glycanases are associated with the tail structure of many tailed phages. Phage ФMR11 is a long-tailed phage that targets Staphylococcus aureus, and protein Gp61, which has glycoside hydrolase activity, is located on the tail of this phage [18]. Phage P22 is a short-tailed phage, and its tail spike protein, Gp26, has endorhamnosidase activity, which can specifically hydrolyze the O-antigen polysaccharide of Salmonella typhimurium [19]. K5 lyase A is also a tail spike protein, and it is encoded by the coliphage K5A, which is capable of degrading K5 exopolysaccharides with the repeating unit [-4)-GlcA-(1,4)-GlcNAc(1-] [20]. Here, Gp190 was also identified on the tail of phage PF-CE2. Therefore, according to the above analysis, there is a strong possibility that Gp190 of phage PF-CE2 is capable of expressing bacteriophage-borne glycanase. Except for the Cronobacter phage vB CsaM leN, all of the phages contained a sequence homologous to Gp190 (> 92% identity, with 100% coverage), suggesting that they may also express bacteriophage-borne glycanase. Regrettably, there have been few studies regarding the glycanases of C. sakazakii phages, so this study is beneficial for exploring which gene encodes glycanase and performing heterologous expression of this enzyme.
For further research and application of Gp190, several online tools were used to study its structure. The primary structure of Gp190 was analyzed using the ProtParam tool (https://web.expasy.org/protparam/) [21], which showed that Gp190 is 1761 bp long and encodes a protein of 586 amino acids. The molecular formula is C2821H4439N797O895S17, with a molecular weight of 64385.10 Da, a theoretical isoelectric point of 5.01, and a liposolubility index of 77.65. The instability index of Gp190 was predicted to be 29.77, which indicated that the protein is stable. The Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) was utilized to predict the secondary structure of Gp190 [22], showing that Gp190 contains 14% alpha helices and 35% beta strands. ProtScale (https://web.expasy.org/protscale/) was used to predict the hydrophilicity and hydrophobicity of the Gp190 protein [21], revealing that hydrophilic amino acids account for 65% of the total, suggesting that Gp190 of phage PF-CE2 may be a soluble protein. No transmembrane domain was identified in Gp190 using the TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Additionally, Gp190 was predicted to be a nonsecreted protein. The SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP-4.1/), and PrediSi (http://www.predisi.de/predisi/index.html) software were used to predict whether Gp190 has a signal peptide, but no signal peptide was identified by either tool.
Although most proteins encoded by phage PF-CE2 were found to be similar to those of the above-mentioned Cronobacter phages, there were still slight differences between them. For instance, the genes Gp202 and Gp262 were found to be unique to phage PF-CE2 (Fig. 1). According to analysis using HHPred software, Gp202 had 96.23% probability of encoding an HNH endonuclease, and the protein encoded by Gp202 was 89% identical to the mobile endonuclease, MobE, encoded by Escherichia virus RB43. HNH endonuclease is important in the life cycle of bacteriophages. Most HNH endonucleases have DNA nicking activity, and MobE is believed to promote the movement of homing endonuclease I-TevIII by cutting the specific non-coding region of the gene encoding the small subunits of aerobic ribonucleotide reductase (nrdB), which is conducive to the inheritance of homing endonuclease I-TevIII in the offspring phages [23, 24]. The protein encoded by Gp202 may be of importance for the reproduction and infectivity of phage PF-CE2. Gp261, Gp262, and Gp263 were all predicted to be related to the synthesis of phage tail fibers. The probability of Gp262 encoding long tail fibers was 98.25% according to HHPred software analysis. Enterobacteria phage T4 (NC_00866), also belonging to the subfamily Tevenvirinae of the family Myoviridae, contains both long and short tail fibers in its tail, which are responsible for recognition of and binding to the host cell. The long tail fiber of phage T4 recognizes outer membrane protein C or the lipopolysaccharides of host bacteria and is responsible for the initial and reversible attachment of the virion [25]. Therefore, Gp262 may be beneficial for phage PF-CE2 to recognize and adsorb to its host.
Previous studies have used the DNA polymerase and short tail fiber protein sequences to determine the phylogenetic relationships of several phages [26, 27]. Phylogenetic trees were made by the neighbor-joining method using MEGA 7.0 [28] and are shown in Supplementary Fig. S1. The tree based on the DNA polymerase shows that phage PF-CE2 grouped with the Cronobacter phage vB CsaM leB, suggesting a close relationship between these phages. In a tree based on the short tail fiber protein, phage PF-CE2 was most similar to the Citrobacter phage Margaery, with a bootstrap value of 98%, and both grouped in a larger cluster that included Cronobacter phages vB CsaM leE and vB CsaM leB. The Bacterial and Archaeal Viruses Subcommittee (BAVS) of the ICTV has specified that the difference between two viruses belonging to the same species should be less than 5% at the nucleotide level. Recently, average nucleotide identity (ANI) has been used to assess the genetic relationships among species at the genome level [29,30,31]. ANI values of genus and species demarcation boundaries have a mean of 73.98% and 95%, respectively [30, 31], and the ANI values based on more than 50% coverage of the genome are considered credible [32]. Here, we used JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/) to estimate the ANI values between the phage PF-CE2 genome and other phage genomes [33]. When compared with phage genomes of 14 members of the subfamily Tevenvirinae of the family Myoviridae, as shown in Fig. 2A and Supplementary Table S5, phage PF-CE2 shared 88.65% genome ANI with Escherichia phage RB16 (genus Pseudotevenvirus, HM134276), which suggests that phage PF-CE2 belongs to the genus Pseudotevenvirus. As shown in Fig. 2B and Supplementary Table S6, phage PF-CE2 and the eight phages mentioned above share < 95% genome ANI with Escherichia phage RB16 but share > 95% genome ANI with each other. This indicates that the nine phages could be assigned to a new species in the genus Pseudotevenvirus of the subfamily Tevenvirinae in the family Myoviridae, and we propose the name "Cronobacter virus PF-CE2" for this new species.
Heat map showing the average nucleotide sequence identity values determined for phage genomes. (A) The PF-CE2 phage genome was compared with 14 phage genomes from members of the subfamily Tevenvirinae of the family Myoviridae, including Acinetobacter_phage_133 (subfamily Tevenvirinae, NC_015250); Aeromonas phage 65 (subfamily Tevenvirinae, NC_015251); Aeromonas phage Aeh1 (subfamily Tevenvirinae, NC_005260); Enterobacteria_phage_Bp7 (genus Dhakavirus, NC_019500); Escherichia phage RB16 (genus Pseudotevenvirus, HM134276); Enterobacteria_phage_vB_EcoM_VR20 (genus Gaprivervirus, NC_028894); Salmonella_phage_Melville (genus Gelderlandvirus, NC_042044); Klebsiella_phage_JD18 (genus Jiaodavirus, NC_028686); Enterobacter_phage_PG7 (genus Karamvirus, NC_023561); Escherichia_phage_ECD7 (genus Krischvirus, NC_041936); Citrobacter_phage_CF1_ERZ-2017 (genus Moonvirus, NC_042067); Escherichia_phage_APCEc01 (genus Mosigvirus, NC_029091); Vibrio_phage_KVP40 (genus Schizotequatrovirus, NC_005083); Enterobacter_phage_phiEap-3 (genus Slopekvirus, NC_041980). (B) The genomes of phage PF-CE2 and eight phages showing a high percentage of sequence similarity to PF-CE2 were compared with the genome of Escherichia phage RB16
In conclusion, we determined the genome sequence of phage PF-CE2, belonging to the subfamily Tevenvirinae of the family Myoviridae. This finding is beneficial for identifying the gene encoding the bacteriophage-borne glycanase of the C. sakazakii phage PF-CE2 and for understanding the mechanism of infection of phage PF-CE2 in host bacteria containing FcEPSs, which could facilitate the wide application of bacteriophage-borne glycanases in the preparation of fucosylated oligosaccharides. Moreover, we propose the establishment of a new species named "Cronobacter virus PF-CE2" and modification of the taxonomic status of eight related phages.
References
Endersen L, Buttimer C, Nevin E, Coffey A, Neve H, Oliveira H, Lavigne R, O’Mahony J (2017) Investigating the biocontrol and anti-biofilm potential of a three phage cocktail against Cronobacter sakazakii in different brands of infant formula. Int J Food Microbiol 253:1–11
Friedemann M (2007) Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int J Food Microbiol 116:1–10
Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S (2006) Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin Infect Dis 42(7):996–1002
Healy B, Cooney S, O’Brien S, Iversen C, Whyte P, Nally J, Callanan JJ, Fanning S (2010) Cronobacter (Enterobacter sakazakii): an opportunistic foodborne pathogen. Foodborne Pathog Dis 7(4):339–350
McMinn A, Liang Y, Wang M (2020) Minireview: The role of viruses in marine photosynthetic biofilms. Mar Life Sci Technol 2:203–208
Zeng HY, He WJ, Li CS, Zhang JM, Li N, Ding Y, Xue L, Chen MT, Wu HM, Wu QP (2019) Complete genome analysis of a novel phage GW1 lysing Cronobacter. Arch Virol 164(2):625–628
Vanhooren PT, Vandamme EJ (1999) L-fucose: occurrence, physiological role, chemical, enzymatic and microbial synthesis. J Chem Technol Biot 74(6):479–497
Elsässer-Beile U, Friebolin H, Stirm S (1978) Primary structure of Klebsiella serotype 6 capsular polyssacharide. Carbohydr Res 65(2):245–249
Xiao MS, Fu XD, Wei XY, Chi YZ, Gao W, Yu Y, Liu ZM, Zhu CL, Mou HJ (2021) Structural characterization of fucose-containing disaccharides prepared from exopolysaccharides of Enterobacter sakazakii. Carbohydr Polym 252:117139
Mcdermott JR, Shao QY, O’Leary C, Kongari R, Liu M (2019) Complete Genome Sequence of Citrobacter freundii Myophage Maroon. Microbiol Resour Announc 8(43):e01426-e1514
Abbasifar R, Kropinski AM, Sabour PM, Ackermann HW, Lingohr EJ, Griffiths MW (2012) Complete genome sequence of Cronobacter sakazakii bacteriophage vB_CsaM_GAP161. J Virol 86(24):13806–13807
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Alikhan NF, Petty NK, Zakour B, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402
Shang AQ, Liu Y, Wang JL, Mo ZL, Li GY, Mou HJ (2015) Complete nucleotide sequence of Klebsiella phage P13 and prediction of an EPS depolymerase gene. Virus Genes 50(1):118–128
Kanamaru S, Ishiwata Y, Suzuki T, Rossmann MG, Arisaka F (2005) Control of bacteriophage T4 tail lysozyme activity during the infection process. J Mol Biol 346(4):1013–1020
Yokoi N, Inaba H, Terauchi M, Stieg AZ, Sanghamitra NJM, Koshiyama T, Yutani K, Kanamaru S, Arisaka F, Hikage T (2010) Construction of robust bio-nanotubes using the controlled self-assembly of component proteins of bacteriophage T4. Small 6(17):1873–1879
Rashel M, Uchiyama J, Takemura I, Hoshiba H, Ujihara T, Takatsuji H, Honke K, Matsuzaki S (2008) Tail-associated structural protein Gp61 of Staphylococcus aureus phage phi MRU has bifunctional lytic activity. FEMS Microbiol Lett 284(1):9–16
Andres D, Hanke C, Baxa U, Seul A, Seckler R (2010) Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J Biol Chem 285(47):36768–36775
Thompson JE, Pourhossein M, Waterhouse A, Hudson T, Goldrick M, Derrick JP, Roberts IS (2010) The K5 lyase KflA combines a viral tail spike structure with a bacterial polysaccharide lyase mechanism. J Biol Chem 285(31):23963–23969
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy Server. In: Walker JM (ed) The proteomics protocols handbook. Springer protocols handbooks, Humana Press, pp 571–607
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Zhang L, Xu D, Huang Y, Zhu X, Rui M, Wan T (2017) Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease. Sci Rep 7:42542
Wilson GW, Edgell D (2009) Phage T4 mobE promotes trans homing of the defunct homing endonuclease I-TevIII. Nucleic Acids Res 37:7110–7123
Meritxell G, Mikiyoshi N, Sara A, Shuji K, Mark VR (2017) Crystallization of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34. Viruses 9(7):970
Sváb D, Falgenhauer L, Rohde M, Szabó J, Chakraborty T, Tóth I (2018) Identification and characterization of T5-like bacteriophages representing two novel subgroups from food products. Front Microbiol 9:202
Senevirathne A, Ghosh K, Roh E, Kim KP (2017) Complete genome sequence analysis of a novel Staphylococcus phage StAP1 and proposal of a new species in the genus Silviavirus. Arch Virol 162(7):2145–2148
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Pacífico C, Hilbert M, Sofka D, Dinhopl N, Hilbertet F (2019) Natural occurrence of Escherichia coli-infecting bacteriophages in clinical samples. Front Microbiol 10:2484
Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2018) High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114
Barco RA, Garrity GM, Scott JJ, Amend JP, Nealson KH, Emersonet D (2020) A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio 11(1):e02475-19
Paepe MD, Hutinet G, Son O, Amarir-Bouhram J, Schbath S, Petit MA, Casadesús J (2014) Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of rad52-like recombinases. PLoS Genet 10(3):e1004181
Richter M, Rosselló-Móra R, Ckner F, Peplies J (2015) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32(6):929–931
Funding
The authors are grateful for financial support from the National Natural Science Found1ation of China (31872893).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have declared no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Nucleotide sequence accession number
The genome sequence of phage PF-CE2 was deposited in the GenBank database under the accession number MW629017.
Additional information
Communicated by Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, M., Ren, X., Yu, Y. et al. Genome sequence analysis of Cronobacter phage PF-CE2 and proposal of a new species in the genus Pseudotevenvirus. Arch Virol 166, 3467–3472 (2021). https://doi.org/10.1007/s00705-021-05255-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05255-z