Abstract
Classical swine fever (CSF), which is caused by classical swine fever virus (CSFV), is a highly contagious disease of pigs. CSFV is genetically and serologically related to bovine viral diarrhea virus (BVDV), a ruminant pestivirus. However, currently available ELISAs based on the full-length E2 protein of CSFV cannot discriminate anti-CSFV from anti-BVDV antibodies. In this study, a truncated CSFV E2 protein (amino acids 690 to 879) covering antigenic domains B/C/D/A (E2B/C/D/A) was designed based on homologous modeling according to the crystal structure of the BVDV E2 protein. The E2B/C/D/A protein was expressed in CHO cells adapted to serum-free suspension culture, and an indirect ELISA (iELISA) was established based on the recombinant protein. No serological cross-reaction was observed for anti-BVDV sera in the iELISA. When testing 282 swine serum samples, the iELISA displayed a high sensitivity (119/127, 93.7%) and specificity (143/155, 92.3%), with an agreement of 92.9% (262/282) and 92.2% (260/282) with virus neutralization test and the IDEXX CSFV blocking ELISA, respectively. Taken together, the newly developed iELISA is highly specific and sensitive and able to differentiate anti-CSFV from anti-BVDV antibodies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Classical swine fever (CSF) is a highly contagious disease of domestic pigs and wild boars, which leads to huge economic losses to the pig industry worldwide. CSF is caused by classical swine fever virus (CSFV), which is taxonomically classified within the Pestivirus genus and Flaviviridae family. Other classifiable members of the Pestivirus genus include bovine viral diarrhea virus (BVDV), border disease virus (BDV), and a growing number of unclassified and so-called atypical pestiviruses (HoBi-like viruses, Bungowannah virus, and atypical porcine pestivirus, etc.) [1, 2]. CSFV is an enveloped virus with a positive-stranded RNA genome of approximately 12.3 kb. The genome contains a single large open reading frame (ORF) encoding a polyprotein of approximately 3,900 amino acids (aa), which is further processed into four structural proteins (C, Erns, E1, and E2) and eight nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) by virus-encoded and cellular proteases [3].
The glycoprotein E2 is anchored to the surface of the virion and can induce neutralizing antibodies (NAbs) upon CSFV infection or vaccination with E2 derived immunogens [4, 5]. The epitope of the CSFV E2 protein has been mapped based on neutralization-escape mutations and competitive antibody binding assays and four distinct antigenic domains have been identified in the order of B/C/D/A located in the N-terminal half of E2. All four domains constitute two independent antigenic units, B/C and D/A linked by disulfide bonds, respectively [6,7,8,9]. Domains B/C have been identified to be responsible for antigenic specificity among various CSFV strains, and domains D/A are relatively conserved [10]. The linear epitope 995YYEP998 is highly conserved among pestiviruses [11], while 829TAVSPTTLR837 is specific to CSFV [12].
BVDV, a ruminant pestivirus, not only infects cattle, but also pigs [13]. Pigs can be infected with BVDV asymptomatically [14]. However, BVDV can also cause CSF-like clinical signs in pigs, including hyperthermia, conjunctivitis, diarrhea and hemorrhages [14]. Serological cross-reactions between BVDV and CSFV have been reported [15, 16], due to a similar genome organization and high homology among pestiviruses. Increasing numbers of BVDV infections in pigs have been documented in many countries, with a seroprevalence of 23-34% in China [17], 3-40% in Austria and Germany [13, 14], 15-20% in Holland [18], and 2-6% in Norway, Denmark and Ireland [19,20,21]. Considering the prevalence of BVDV in pig populations and the wide use of BVDV-contaminated bovine sera for production of swine vaccines [14, 22], this cross-reactivity is a major bottleneck for accurate diagnosis of CSF. However, ELISAs based on the full-length E2 protein of CSFV cannot discriminate anti-CSFV from anti-BVDV antibodies. Moreover, currently available commercial ELISA kits are expensive, which limits their wide application. Thus, a serological diagnostic assay with the potential to effectively distinguish CSFV from BVDV is urgently needed.
The crystal structure of the BVDV E2 has recently been resolved, in which two Ig-like domains form a unique protein architecture that contains an elongated β-stranded domain with a new fold at the C-terminus [23]. Researchers have also attempted to map the antigenic regions of CSFV E2 in the structure of BVDV E2 according to the proposed antigenic domain of CSFV [23]. Here, we predicted the structure of CSFV E2 using homologous modeling techniques, based on the crystal structure of the BVDV E2 and designed a truncated CSFV E2 protein (aa 690–879) covering antigenic domains B/C/D/A (E2B/C/D/A). The E2B/C/D/A protein was expressed in a mammalian expression system, and an indirect enzyme-linked immunosorbent assay based on the recombinant protein was established (E2B/C/D/A-based iELISA). This assay was evaluated in comparison with virus neutralization test (VNT) and the IDEXX CSFV blocking ELISA.
Materials and methods
Cells, viruses, and antisera
Both porcine kidney (PK-15) cells and human embryonic kidney (HEK)-293T cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS, Sigma-Aldrich, USA) and maintained at 37°C in 5% CO2. Chinese hamster ovary (CHO) cells were cultured in DME/F-12 (GE Healthcare, USA) containing 5% FBS and maintained at 37°C in 5% CO2. The CSFV Shimen strain was propagated in PK-15 cells and recombinant lentiviruses were grown in HEK-293T cells.
Two hundred eighty-two swine serum samples, which have been determined to be positive or negative against the CSFV Shimen strain by VNT, were used in this study (Table S1). VNT was performed according to the EU Diagnostic Manual for CSF Diagnosis [24]. Briefly, the diluted sera (inactivated for 30 min at 56°C) were incubated with an equal volume of a viral suspension that contains 200 TCID50 CSFV Shimen strain for 1 h at 37°C. Thereafter the serum/virus mixture was added to confluent monolayers of PK-15 cells in 96-well cell culture plates (Corning, USA) and incubated at 37°C for 72 h. The CSFV-specific neutralizing antibody (NAb) titers were expressed as the reciprocal of the highest serum dilution that inhibited the infection of PK-15 cells in 50% of the culture wells. Seven sera was collected from pigs experimentally infected with BVDV NADL, 22146, 1764/4, Oregon C24V or YS09 strain at 59–179 days post-infection (dpi) (Table 1). The NAb titers of BVDV-infected swine sera against BVDV or CSFV were determined by VNT as described previously [24] in MDBK cells for BVDV and in PK-15 cells for CSFV (Table 1). The 15 other serum samples are from pigs experimentally infected with porcine reproductive and respiratory syndrome virus (PRRSV) (5), pseudorabies virus (PRV) (4) or porcine circovirus type 2 (PCV2) (6).
Structure prediction and design of truncated CSFV E2 based on that of BVDV E2
The E2 sequence of the CSFV Shimen strain was obtained from the National Center for Biological Information (NCBI) database under accession number AY775178.2. The sequence was then submitted to the ExPASy modelling server (http://swissmodel.expasy.org/workspace/) to build the three-dimensional (3D) model. Template selection, alignment and model building were done automatically by the server. The CSFV E2 protein was truncated based on the predicted 3D model.
Construction of recombinant plasmid expressing truncated CSFV E2
Total RNA was extracted from the supernatants of CSFV-infected PK-15 cells using the QIAamp® Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s manual and reverse transcribed to cDNA as previously described [25]. The E2B/C/D/A-coding region with a signal peptide was amplified by PCR from the cDNA using primers P1 (5'- CGGAATTCGCCACCATGGTATTAAGAGGACAGATCGTGC-3') and P2 (5'- ATCTCGAGTTACTTCTCGAACTGGGGGTGGGACCATGCTGAATGATGATGATGATGATGCCCTTTCACACATGTCCAGTT-3') with the introduced EcoRI and XhoI sites and the coding sequences for the C-terminal Strep and 6×His tags. The PCR product was cloned into the lentiviral expression vector pLVX-IRES-ZsGreen1 (Clontech). The resulting plasmid was further confirmed by sequencing and designated as pLVX-E2B/C/D/A.
Establishment of a cell line stably expressing the CSFV E2B/C/D/A protein
To generate a cell line stably expressing E2B/C/D/A, HEK-293T cells cultured in a 10-cm cell culture dish were transfected with 21 μg of pLVX-E2B/C/D/A, together with 14 μg of the packaging plasmid psPAX2 (Addgene) and 7 μg of the shuttle plasmid pMD2.G (Addgene). Six hours post-transfection (hpt), the transfection medium was replaced with DMEM supplemented with 10% FBS and further incubated for 48 h. The recombinant lentiviruses were harvested by collecting the supernatants of the transfected cells, and subsequently transduced into CHO cells. The clonal isolation of recombinant cells expressing the E2 protein was performed and confirmed by ELISA and Western blotting. The recombinant cells were further adapted to and grown in suspension culture (CHOGrow CD2 Serum-Free Medium, BasalMedia, China). The supernatants from the cell culture containing the E2B/C/D/A protein were purified with StrepTactin Sepharose according to the manufacturer’s guidelines (GE Healthcare, USA).
Western blotting
The recombinant protein was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membrane by using a semi-dry transfer cell (Bio-Rad, USA) according to the manufacturer’s manual. The membrane was treated sequentially with blocking solution (PBS containing 5% skim milk), monoclonal antibody MAb WH303 against CSFV E2 (1,000-fold dilution) and IRDye800CW goat anti-mouse IgG (Licor, Odyssey, USA; 10,000-fold dilution). Finally, the fluorescence signal was detected by an Infrared Fluorescence Imaging System (Licor, Odyssey).
Development and optimization of the E2B/C/D/A-based iELISA
To establish an efficient iELISA, the concentrations of component reagents for the iELISA and different assay parameters were optimized. The optimal concentrations of the coating antigen (purified E2B/C/D/A) and dilutions of sera and secondary antibodies [horseradish peroxidase (HRP)-conjugated rabbit anti-pig IgG] (Sigma-Aldrich, USA) were determined according to the checkerboard titration method [26].
Ninety-six-well ELISA plates were coated with 100 μL of the purified recombinant protein (2.5 μg/mL) diluted in carbonate–bicarbonate buffer (pH = 9.6) and incubated at 4°C overnight. The coated plates were washed with PBS containing 0.05% Tween-20 (PBST) and blocked with 5% skim milk in PBS for 1 h at 37°C. Subsequently, serum samples were diluted in PBST containing 5% skim milk (1:100) and incubated at 37°C for 1 h, and then the plates were washed with PBST. Thereafter, the plates were incubated with 100 μL of the HRP-conjugated rabbit anti-pig IgG (Sigma-Aldrich) diluted in PBST containing 5% skim milk (1:20,000) for 1 h at 37°C. After washing with PBST for five times, the bound antibodies were detected with TMB (100 μL/well) (Sigma-Aldrich, USA) at room temperature. The reaction was stopped using 100 μL of 2 M H2SO4 in each well and the optical density (OD) values were examined at 450 nm in a microtiter plate reader (Biotek, USA).
Evaluation of the E2B/C/D/A-based iELISA
The specificity and sensitivity of the E2B/C/D/A-based iELISA was tested by using 282 swine sera, confirmed by VNT. The cutoff value of the iELISA was determined by SigmaPlot software version 11.0 (Systat Software, Inc.). In addition, the sera from the pigs experimentally infected with BVDV, PRRSV, PRV, or PCV2 were used to evaluate cross-reactivity within the assay. The sensitivity of the assay was evaluated using four serum samples with different NAb titers.
Comparison of the E2B/C/D/A-based iELISA with VNT and the IDEXX CSFV blocking ELISA
The performance of the E2B/C/D/A-based iELISA was compared with VNT and the IDEXX CSFV blocking ELISA (IDEXX, USA) assays using 282 swine serum samples. VNT was performed as described previously [24] while the blocking ELISA was performed according to the manufacturer’s guidelines. The sensitivity and specificity were calculated by using the “Diagnostic or Screening Test Evaluation” online software (http://www.openepi.com).
Results
Putative 3D structure of the CSFV E2 protein
The E2 sequence of CSFV Shimen strain (aa 690–1,062) was submitted to the Swiss model server to search for suitable templates. The structure of the BVDV E2 [Protein Data Bank (PDB) code 4jnt.1.B] [23] was identified as the best template candidate with 61% sequence similarity to the E2 sequence of the CSFV Shimen strain. Based on the 3D model of the CSFV E2 protein (Fig. 1B), generated according to the crystal structure of the BVDV E2 (Fig. 1A), a truncated E2 protein covering the antigenic domains B/C/D/A (E2B/C/D/A, aa 690–879) was designed (Fig. 1B), removing the elongated β-stranded domain, which contains a linear epitope (995YYEP998) highly conserved among pestiviruses [11].
The crystal structure of the E2 protein of bovine viral diarrhea virus (BVDV) (A), and the putative structure of the E2 protein of classical swine fever virus (CSFV) (B). Homology modeling of the CSFV E2 protein based on the crystal structure of BVDV E2. The truncated version of the CSFV E2 protein (aa 690–879) covers the antigenic domains B, C, D, and A. Domains B/C are shown in red, domains D/A in yellow, and the elongated β-stranded domain in blue
Expression and identification of the E2B/C/D/A protein
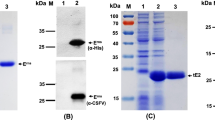
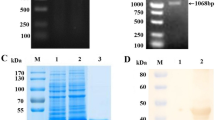
The lentiviral expression vector pLVX-IRES-ZsGreen1 allows the simultaneous expression of a protein of interest and green fluorescent protein (ZsGreen1) in mammalian cells. Thus, the expression of the E2B/C/D/A protein in CHO cells (CHO-E2B/C/D/A) could be monitored using ZsGreen1 as an indicator (Fig. 2A). The cells obtained by a clonal isolation process were adapted successfully to a protein-free and serum-free suspension culture. Western blotting analysis of the CHO-E2B/C/D/A cells showed a specific band between 25 kDa and 35 kDa which was recognized by the anti-E2 MAb WH303 [27] (Fig. 2B).
Expression of the E2B/C/D/A protein in CHO cells. (A) CHO cells were transduced with the recombinant lentiviruses packaged with the lentiviral expression vector pLVX-IRES-ZsGreen1. The expression of the E2B/C/D/A protein in the resulting recombinant CHO (CHO-E2B/C/D/A) cells was monitored using green fluorescent protein (ZsGreen1) as an indicator, and mock-transduced CHO cells were used as a negative control. (B) Western blotting analysis of the E2B/C/D/A protein using monoclonal antibody (MAb) WH303 against the CSFV E2. Lane M, protein marker; lane 1, mock-transduced CHO cells; lane 2, purified E2B/C/D/A protein
Optimization of the E2B/C/D/A-based iELISA
The iELISA parameters were optimized with the concentration of coating antigen being 2.5 μg/mL and the dilution of serum samples and secondary antibody dilution being 1:100 and 1:20,000, respectively (Fig. 3). Following these optimal conditions, a total of 282 swine serum samples were tested by the assay and accordingly the cutoff absorbance value was determined to be 0.2. Samples with OD values greater than or equal to the cutoff value were considered positive.
Specificity of the E2B/C/D/A-based iELISA
To validate the potential of the E2B/C/D/A-based iELISA for detecting specific antibodies against CSFV, anti-sera specific for BVDV, PRRSV, PRV or PCV2 were used to determine the specificity of the assay. As expected, the absorbance values of all these sera were lower than the cutoff value (Fig. 4).
Sensitivity of the E2B/C/D/A-based iELISA
The sensitivity of the E2B/C/D/A-based iELISA was evaluated using serum samples with different NAb titers. As shown in Fig. 5, the iELISA titers of the CSFV antibody positive sera were equal to or greater than the corresponding NAb titers.
Performance of the E2B/C/D/A-based iELISA in serodiagnosis
A total of 282 serum samples were used to evaluate the potential of the E2B/C/D/A-based iELISA, in comparison with VNT and the IDEXX CSFV blocking ELISA. The iELISA displayed a high sensitivity (119/127, 93.7%) [95% confidence interval (CI): 88.1–96.7%] and specificity (143/155, 92.3%) (95% CI: 87.0–95.5%) comparable to VNT, and the agreement rate was 92.9% (262/282) and 92.2% (260/282) with VNT and the blocking ELISA, respectively (Table 2).
Discussion
CSF poses a serious threat to the global pig industry, especially in China [1, 28]. Rapid and accurate serological diagnosis is a crucial step for evaluation of immunization effects and efficient control of CSF. However, since pestiviruses share similar genomic organization and antigenic structures, serological cross-reactivity can occur between CSFV and BVDV, a ruminant pestivirus capable of infecting pigs [14, 29]. Cross-reaction between antibodies against BVDV and CSFV has been reported [16, 30]. The practical implications of this is that the presence of ruminant pestivirus antibodies in swine often causes false positive reactions in serological surveys for CSFV, which raises problems during epidemiological surveillance for CSFV and in CSFV eradication campaigns. The currently available commercial ELISA tests for CSFV are mostly based on the entire E2 protein, which can lead to false positive results [16, 31]. Moreover, the commercial ELISA kits are not cost-effective, which limits their application in countries with high pig densities. Accordingly, CSFV-specific serological assays with the potential to effectively differentiate CSFV antibodies from BVDV antibodies are urgently needed.
At present, the E2 protein of CSFV has been extensively studied and four independent antigenic domains have been identified in the N-terminus in the order of B, C, D and A. Both linear [4, 11, 12, 32, 33] and conformational epitopes [9, 34,35,36,37] have been defined in the N-terminal half of E2. Homology modeling techniques have been widely used to predict 3D protein models based on the experimentally determined structures of homologous proteins. Fortunately, the crystal structure of the BVDV E2 protein has recently been resolved [23], making it possible to predict the structure of the CSFV E2 protein by homologous modeling, as BVDV shares a similar genomic organization with CSFV. A truncated E2 (E2B/C/D/A) covering the antigenic domains B/C/D/A of E2 (aa 690–879) was designed in which the elongated β-stranded domain was deleted. As expected, the E2B/C/D/A-based iELISA was highly specific for CSFV, as no cross-reaction was observed with BVDV in the assay. The successful elimination of cross-reaction with anti-BVDV antibodies is likely due to the deletion of the highly conserved linear epitope (995YYEP998) found in the E2 protein of pestiviruses.
Mammalian cell expression systems are widely used in antigen preparation, since they combine the advantages of ease of use, low-cost and high-yield. Here, we chose CHO cells for the expression of the recombinant protein. CHO cells are more frequently used for stable transfection, since the cells are capable of adapting to and growing in suspension culture, which is ideal for large-scale production. Furthermore, CHO cells can grow in a serum-free and chemically defined medium, which can ensure reproducibility between different batches of cell culture [38]. The truncated E2 was constitutively expressed in CHO cells and readily adapted to suspension culture with serum-free and chemically defined medium, indicating the prospect of large-scale production of this protein. Next, an iELISA, based on the E2B/C/D/A protein, was developed (with optimized conditions) and validated using a panel of serum samples with expected results being achieved.
Generally, VNT is the gold standard for detection of anti-CSFV antibodies, as according to the World Organisation for Animal Health (OIE). However, this method is time-consuming, labor-intensive and inappropriate for large-scale screening. To evaluate the clinical application potential of the E2B/C/D/A-based iELISA, a total of 282 serum samples were tested and compared with VNT and the IDEXX CSFV blocking ELISA. The iELISA showed a high agreement with these two assays, indicating the high performance of the newly developed assay. The E2B/C/D/A-based iELISA has a potential to be used for rapid and efficient screening of a large number of samples, especially for assessment of the immune status of pigs following vaccination in countries with a high pig density.
Chang et al. have analyzed antigenic domains through expressing a series of truncated E2 proteins [10]; however, these have not been put into use. In this study, we established a CHO cell line continuously expressing the truncated E2 protein and developed a cost-effective iELISA method, which might be applicable for large-scale surveillance in the field. Further work is needed to validate the applicability of the iELISA for various samples, such as blood and ear notches.
Conclusion
In summary, this is the first report to show that the truncated E2 protein of CSFV can be ‘structurally designed’ by homologous modeling based on the crystal structure of the BVDV E2. An E2B/C/D/A-based iELISA established in this study was highly specific and sensitive and has the potential for specific serological surveillance of CSF. The next step is to facilitate the development of an ELISA kit for large-scale clinical detection of anti-CSFV antibodies at low cost.
References
Luo Y, Li S, Sun Y, Qiu HJ (2014) Classical swine fever in China: a minireview. Vet Microbiol 172:1–6
Blome S, Staubach C, Henke J, Carlson J, Beer M (2017) Classical swine fever-an updated review. Viruses 9:86
Meyers G, Thiel HJ (1996) Molecular characterization of pestiviruses. Adv Virus Res 47:53–118
Zhang F, Yu M, Weiland E, Morrissy C, Zhang N, Westbury H, Wang LF (2006) Characterization of epitopes for neutralizing monoclonal antibodies to classical swine fever virus E2 and Erns using phage-displayed random peptide library. Arch Virol 151:37–54
Qi Y, Zhang BQ, Shen Z, Chen YH (2009) Candidate vaccine focused on a classical Swine fever virus epitope induced antibodies with neutralizing activity. Viral Immunol 22:205–213
Wensvoort G, Terpstra C, Boonstra J, Bloemraad M, Van Zaane D (1986) Production of monoclonal antibodies against swine fever virus and their use in laboratory diagnosis. Vet Microbiol 12:101–108
Wensvoort G (1989) Topographical and functional mapping of epitopes on hog cholera virus with monoclonal antibodies. J Gen Virol 70:2865–2876
Wensvoort G, Boonstra J, Bodzinga BG (1990) Immunoaffinity purification and characterization of the envelope protein E1 of hog cholera virus. J Gen Virol 71:531–540
van Rijn PA, Miedema GK, Wensvoort G, van Gennip HG, Moormann RJ (1994) Antigenic structure of envelope glycoprotein E1 of hog cholera virus. J Virol 68:3934–3942
Chang CY, Huang CC, Lin YJ, Deng MC, Chen HC, Tsai CH, Chang WM, Wang FI (2010) Antigenic domains analysis of classical swine fever virus E2 glycoprotein by mutagenesis and conformation-dependent monoclonal antibodies. Virus Res 149:183–189
Yu M, Wang LF, Shiell BJ, Morrissy CJ, Westbury HA (1996) Fine mapping of a C-terminal linear epitope highly conserved among the major envelope glycoprotein E2 (gp51 to gp54) of different pestiviruses. Virology 222:289–292
Lin M, Lin F, Mallory M, Clavijo A (2000) Deletions of structural glycoprotein E2 of classical swine fever virus strain alfort/187 resolve a linear epitope of monoclonal antibody WH303 and the minimal N-terminal domain essential for binding immunoglobulin G antibodies of a pig hyperimmune serum. J Virol 74:11619–11625
Liess B, Moennig V (1990) Ruminant pestivirus infection in pigs. Rev Sci Tech 9:151–161
Tao J, Liao J, Wang Y, Zhang X, Wang J, Zhu G (2013) Bovine viral diarrhea virus (BVDV) infections in pigs. Vet Microbiol 165:185–189
Liu L, Hoffmann B, Baule C, Beer M, Belák S, Widén F (2009) Two real-time RT-PCR assays of classical swine fever virus, developed for the genetic differentiation of naturally infected from vaccinated wild boars. J Virol Methods 159:131–133
Sung JH, Kang ML, Lee WJ, Shin MK, Lim SI, Kim BH, Song JY, Yoo HS (2011) Improved sero-monitoring assay for classical swine fever (CSF) using the recombinant E2 protein of a recent Korean isolate. Res Vet Sci 90:329–335
Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, Zhai SL, Huang L, Shan TL, Zheng H, Wen XT, Tong GZ (2012) High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol 159:490–493
Terpstra C, Wensvoort G (1991) Bovine virus diarrhea virus infections in swine. Tijdschr Diergeneeskd 116:943–948
Løken T, Krogsrud J, Larsen IL (1991) Pestivirus infections in Norway. Serological investigations in cattle, sheep and pigs. Acta Vet Scand 32:27–34
Jensen MH (1985) Screening for neutralizing antibodies against hog cholera- and/or bovine viral diarrhea virus in Danish pigs. Acta Vet Scand 26:72–80
Graham DA, Calvert V, German A, McCullough SJ (2001) Pestiviral infections in sheep and pigs in Northern Ireland. Vet Rec 148:69–72
Wensvoort G, Terpstra C (1988) Bovine viral diarrhoea virus infections in piglets born to sows vaccinated against swine fever with contaminated vaccine. Res Vet Sci 45:143–148
Li Y, Wang J, Kanai R, Modis Y (2013) Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc Natl Acad Sci USA 110:6805–6810
Anonymous (2007) EU Diagnostic Manual for Classical swine fever (CSF) Diagnosis: technical part (Third Draft June 2007). EU and OIE Reference Laboratory for Classical Swine Fever Virus. http://www.tiho-hannover.de/kliniken-institute/institute/institut-fuer-virologie-zentrum-fuer-infektionsmedizin/eu-and-oie-reference-laboratory/downloads/. Accessed 21 Mar 2018
Zhang XJ, Han QY, Sun Y, Zhang X, Qiu HJ (2012) Development of a triplex TaqMan real-time RT-PCR assay for differential detection of wild-type and HCLV vaccine strains of classical swine fever virus and bovine viral diarrhea virus 1. Res Vet Sci 92:512–518
Reed LJ, Muench H (1938) A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497
Edwards S, Sands JJ (1990) Antigenic comparisons of hog cholera virus isolates from Europe, America and Asia using monoclonal antibodies. Dtsch Tierarztl Wochenschr 97:79
Edwards S, Fukusho A, Lefèvre PC, Lipowski A, Pejsak Z, Roehe P, Westergaard J (2000) Classical swine fever: the global situation. Vet Microbiol 73:103–119
Wieringa-Jelsma T, Quak S, Loeffen WL (2006) Limited BVDV transmission and full protection against CSFV transmission in pigs experimentally infected with BVDV type 1b. Vet Microbiol 118:26–36
van Rijn PA (2007) A common neutralizing epitope on envelope glycoprotein E2 of different pestiviruses: implications for improvement of vaccines and diagnostics for classical swine fever (CSF)? Vet Microbiol 125:150–156
Kumar R, Barman NN, Khatoon E, Kumar S (2016) Development of single dilution immunoassay to detect E2 protein specific classical swine fever virus antibody. Vet Immunol Immunopathol 172:50–54
Kortekaas J, Vloet RP, Weerdmeester K, Ketelaar J, van Eijk M, Loeffen WL (2010) Rational design of a classical swine fever C-strain vaccine virus that enables the differentiation between infected and vaccinated animals. J Virol Methods 163:175–185
Peng WP, Hou Q, Xia ZH, Chen D, Li N, Sun Y, Qiu HJ (2008) Identification of a conserved linear B-cell epitope at the N-terminus of the E2 glycoprotein of Classical swine fever virus by phage-displayed random peptide library. Virus Res 135:267–272
Chang CY, Huang CC, Deng MC, Huang YL, Lin YJ, Liu HM, Lin YL, Wang FI (2012) Antigenic mimicking with cysteine-based cyclized peptides reveals a previously unknown antigenic determinant on E2 glycoprotein of classical swine fever virus. Virus Res 163:190–196
van Rijn PA, van Gennip HG, de Meijer EJ, Moormann RJ (1993) Epitope mapping of envelope glycoprotein E1 of hog cholera virus strain Brescia. J Gen Virol 74:2053–2060
van Rijn PA, Bossers A, Wensvoort G, Moormann RJ (1996) Classical swine fever virus (CSFV) envelope glycoprotein E2 containing one structural antigenic unit protects pigs from lethal CSFV challenge. J Gen Virol 77:2737–2745
van Rijn PA, van Gennip RG, de Meijer EJ, Moormann RJ (1992) A preliminary map of epitopes on envelope glycoprotein E1 of HCV strain Brescia. Vet Microbiol 33:221–230
Newman-Tancredi A, Wootton R, Strange PG (1992) High-level stable expression of recombinant 5-HT1A 5-hydroxytryptamine receptors in Chinese hamster ovary cells. Biochem J 285:933–938
Acknowledgements
We thank Prof. Paul Becher at the EU Reference Laboratory for CSF, Hannover, Germany, for providing anti-BVDV sera.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31402194 and 31572540) and Harbin Special Fund for Innovation Talents of Science and Technology (2015RAQYJ064).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no competing interests.
Ethical approval
None of the experiments in this study involved human participants.
Additional information
Handling Editor: Sheela Ramamoorthy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ji, S., Luo, Y., Zhang, T. et al. An improved indirect ELISA for specific detection of antibodies against classical swine fever virus based on structurally designed E2 protein expressed in suspension mammalian cells. Arch Virol 163, 1831–1839 (2018). https://doi.org/10.1007/s00705-018-3809-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3809-7