Abstract
A novel virus, with characteristics of viruses classified within the genus Vitivirus, was identified from a sample of Vitis vinifera cv. Chardonnay in New Zealand. The virus was detected with high throughput sequencing (small RNA and total RNA) and its sequence was confirmed by Sanger sequencing. Its genome is 7507 nt long (excluding the polyA tail) with an organisation similar to that described for other classifiable members of the genus Vitivirus. The closest relative of the virus is grapevine virus E (GVE) with 65% aa identity in ORF1 (65% nt identity) and 63% aa identity in the coat protein (66% nt identity). The relationship with GVE was confirmed with phylogenetic analysis, showing the new virus branching with GVE, Agave tequilina leaf virus and grapevine virus G (GVG). A limited survey revealed the presence of this virus in multiple plants from the same location where the newly described GVG was discovered, and in most cases both viruses were detected as co-infections. The genetic characteristics of this virus suggest it represents an isolate of a new species within the genus Vitivirus and following the current nomenclature, we propose the name “Grapevine virus I”.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Main text
The use of high throughput sequencing (HTS) has had a massive effect on the rate of discovery of previously overlooked obligate parasites. The decrease in technology costs combined with uptake of the methodology by more diagnostic laboratories has resulted in an unprecedented level of detection of novel virus genomes (reviewed in [1]). We reported earlier the identification of a new vitivirus, named grapevine virus G (GVG) from small RNA (sRNA) sequencing and total RNA sequencing [2]. While characterising GVG, we identified, from the same HTS run, a second, related vitivirus. In order to maintain accepted taxonomical consistency we propose to name this new virus isolate grapevine virus I (GVI) and we use this abbreviation hereafter.
A sample of Vitis vinifera cv Chardonnay (VID499 – TK0004) was collected from the New Zealand Winegrowers’ germplasm collection, Lincoln, New Zealand, in November 2016 and total RNA was submitted to RNASeq at the Australian Genome Research Facility. This was analysed on a HiSeq 2500 sequencer after DNase RQ1 treatment alongside the sample VID561 - TK06562 in which the GVG virus was described. Using the bioinformatics pipeline previously described [2] a vitivirus-like sequence of 7439 nt was retrieved from the de novo analysis. In light of this new sequence, we examined the sRNA (sRNA) data obtained previously [2] from the same plant and we found that the contig was mapped by 217532 reads (579 x coverage). The sequence was confirmed by Sanger sequencing. The genome was completed by targeting the 5´ UTR sequence with the SMARTer® RACE 5´/3´ Kit (Clontech Laboratories, Inc. A Takara Bio Company) and the 3´ UTR with an RT-PCR with an oligo(dT) anchored reverse primer. The full genome of 7507 nt, excluding the polyA tail, was deposited in GenBank, as grapevine virus I, under the accession number MF927925. In addition to this virus, the same plant was found to be infected with several viruses including grapevine leafroll-associated virus 3, grapevine virus A, grapevine rupestris stem pitting virus, grapevine rupestris vein feathering virus (MF000326), grapevine redglobe virus, grapevine virus G (MF405924), and the viroids hop stunt viroid and grapevine yellow speckle viroid; these were confirmed from the sRNA data using the YABI Virus Surveillance and Diagnosis (VSD) toolkit [3, 4].
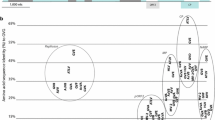
The genome of GVI is comparable to descriptions of vitiviruses [5, 6] with a single positive single-stranded RNA molecule containing five open reading frames (ORFs). ORF1 encodes a 1696 aa polyprotein (nt position 69-5159) that contains the recognised domains of a methyltransferase; helicase; 2OG-Fe(II) oxygenase superfamily (AlkB); and RNA-dependent RNA-Polymerase (RdRp). The closest relative available on GenBank is grapevine virus E (GVE, isolate SA94, GU903012) with 65% aa and nt identity. The second ORF (ORF2) overlaps with ORF1 by 11 nt (nt position 5149-5652) and encodes for a 167 aa putative protein with poor homology to known proteins and no recognised domains, as observed in previously characterised viruses classified within the same genus [6]. The third ORF (ORF3) starts 32 nt downstream of ORF2 and encodes a 264 aa protein (nt position 5685-6479) containing a viral movement protein domain. The movement protein of GVE is its closest relative, with 63% aa identity (65% nt). The next ORF (ORF4) overlaps with ORF3 by 70 nt and encodes a 199 aa protein (nt position 6409-7008) containing the tricho coat super family domain. This protein shares 65% aa identity with the coat protein of agave tequilina leaf virus (ATLV) (68% nt); 63% aa with GVE (66% nt) and 62% aa with GVG (61% nt). ORF5 starts 29 nt downstream of ORF4 and encodes a 121 aa protein (nt position 7038-7403) with a recognised viral nucleic acid binding protein (NABP). This protein shares the greatest degree of conservation with GVE, the closest match available from GenBank (72% aa and 70% nt identity). It is interesting to note that the NABP of Grapevine virus B (GVB) is the second closest match, with 66% aa identity, as none of the other proteins of vitivirus GVB group within the GVE clade (Figure 1).
Schematic representation of the genome organisation of grapevine virus I (GVI). Open reading frames are represented by boxes with the conserved domains italicised. Acronyms used are: the methyl transferase domain (MTR), the helicase domain (HEL), the 2OG-Fe(II) oxygenase domain (AlkB), the RNA-dependant RNA-polymerase domain (RdRp), the movement protein (MP), the coat protein (CP) and the nucleic acid binding protein (NABP). The numbers at the edges of the boxes represent the first and last nucleotide postion of the ORF. The white box with AlkB (GVA) represents the alternative location of this domain for some other members of the vitivirus (GVA clade). Insets represent the Neighbor-Joining trees (1000 bootstrap replicates using the Jukes-Cantor distance model) of the a. replicase polyprotein, b. coat protein and c. AlkB domain of representative members of the genus Vitivirus. Protein alignments (translated from the accession number indicated) were performed with ClustalW (BLOSUM cost matrix with a gap opening cost set at 10, and a gap extension cost at 0.1). Consensus support is shown as a percentage on the branch. Citrus leaf blotch virus, apple chlorotic leaf spot virus and Actinidia virus-1 were used as outgroups for the phylogenetic analysis of the a. replicase b. coat protein and c. AlkB, respectively
A phylogenetic analysis was conducted on the replicase and coat protein genes following a ClustalW aligment (BLOSUM cost matrix with gap opening cost set at 10 and gap extend cost at 0.1), made using the Neighbor-joining method and the Jukes-Cantor genetic distance model. The citrus leaf blotch virus replicase (JN983456) and apple chlorotic leaf spot virus coat protein (CAE52495) were used as outgroups. The replicase phylogenetic tree (Figure 1a) shows GVI branching off the GVE cluster before GVG and ALTV. For the coat protein, although not strongly supported, GVI clusters with GVG and ATLV within the GVE group (Figure 1b).
From a limited survey of old vine accessions located in the same germplasm collection, Lincoln, New Zealand, we identified eight GVI positive vines (including the original VID499-TK0004) from 18 plants tested by sRNA HTS, with 196 to 8209 reads per million mapping to the genome MF927925 (coverage of 92 to 100% of the genome; 17- to 842-fold coverage). Five positive samples were from V. vinifera (Sylvaner, Chardonnay, Dolcetto or Shiraz), one was a Vitis labrusca (Fredonia) and two were interspecific hybrids (Chelois and Pinard). In order to evaluate the establishment of the virus, 58 additional vines (34 from the germplasm and 24 from a commercial vineyard) were tested for the presence of GVI using cDNA synthetized from immunocaptured dsRNA [7], followed by a PCR using the primers GVG-GVI 4595F (TTY TCT CAG AAG ART TAY GAT GAT C) and GVI 5212R (TAT GTT CAG CTC ATG AAG GTG CTC) and Sanger sequencing. Two additional infections were detected from the germplasm but the virus was not detected in the plants sourced from commercial vineyards.
The replicase gene structure of the vitiviruses shows a shift in position of the AlkB domain. AlkB proteins are widely distributed in cellular organisms but are only found in viruses infecting perennial hosts. Their role may involve protection against methylation damage. In the replicase polyprotein, the AlkB domain is found within the helicase domain for GVE, GVG, arracacha virus V (AVV) and GVI, as opposed to other vitiviruses (GVA clade) that have their AlkB domain located upstream of the helicase (Figure 1). This alteration of the genome arrangement supports the hypothesis that GVE and relatives have gained their AlkB domains horizontally, independent from the GVA clade [8]. This explains differences among the sequences of these domains, where GVE, GVG, AVV and GVI cluster together (Figure 1c). Although, AVV is genetically related with members of the GVA clade (Figure 1a and 1b), the position and amino acid sequence of its AlkB domain is related to members of the GVE clade (Figure 1c) suggesting a similar origin (Figure 1c). ATLV is the only vitivirus without an AlkB domain and this absence explains its shorter genome (by about 350 nt), a difference that corresponds to the length of this motif. The phylogenetic analysis of the replicase (Figure 1a) suggests that the virus never incorporated this motif (more basal and thereby older branching). Despite its close relationship with GVE, GVG and GVI, ATLV is not only unique because it lacks the AlkB domain but also because it lacks ORF2. Agave is the only known monocotyledon infected with a vitivirus to date, and does not undergo traditional secondary growth with production of secondary phloem; this may therefore provide a clue for the function of the AlkB domain and ORF2 protein.
The genome of the new virus described herein falls below the threshold for species demarcation within the genus Vitivirus; of 80% aa identity or 72% nt identity for the coat protein and the RdRp [4]. Therefore, we propose that it is considered as a representative isolate of a new species in the genus with the name “Grapevine virus I”.
In the 20-year interval between its establishment (1997) [5] and its review (during the last ICTV update in 2016) the genus Vitivirus has gained five species, from the four original members (Grapevine virus A, Grapevine virus B, Grapevine virus D and Heracleum latent virus) to nine (Grapevine virus E, Grapevine virus F, Actinidia virus A, Actinidia virus B and Mint virus 2) [9,10,11,12]. Eighteen months since that last ICTV release, four new virus isolates that fit the description of viruses classifiable within the genus have been reported or deposited in GenBank: AVV, ATLV (KY190215), GVG and grapevine virus H [2, 13, 14]. In addition, the full genome of GVD was released (MF774336). With the description of GVI, and assuming they are all accepted within the genus in the next ICTV release, the number of approved vitivirus species will increase to 14, eight of which have been described in grapevine.
References:
Roossinck MJ (2017) Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res 239:82–86
Blouin AG, Keenan S, Napier KR, Barrero RA, MacDiarmid RM (2018) Identification of a novel vitivirus from grapevines in New Zealand. Arch Virol 163:281–284
Barrero RA, Napier KR, Cunnington J, Liefting L, Keenan S, Frampton RA, Szabo T, Bulman S, Hunter A, Ward L, Whattam M, Bellgard MI (2017) An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinform 18:26
Hunter AA, Macgregor AB, Szabo TO, Wellington CA, Bellgard MI (2012) Yabi: An online research environment for grid, high performance and cloud computing. Source Code Biol Med 7:1
Adams MJ, Antoniw JF, Bar-Joseph M, Brunt AA, Candresse T, Foster GD, Martelli GP, Milne RG, Zavriev SK, Fauquet CM (2004) The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 149:1045–1060
Minafra A, Mawassi M, Goszczynski D, Saldarelli P (2017) Grapevine vitiviruses. In: Meng B, Martelli G, Golino D, Fuchs M (eds) Grapevine viruses: molecular biology, diagnostics and management. Springer, Cham, pp 229–256
Blouin AG, Ross HA, Hobson-Peters J, O’Brien CA, Warren B, MacDiarmid R (2016) A new virus discovered by immunocapture of double-stranded RNA, a rapid method for virus enrichment in metagenomic studies. Mol Ecol Resour 16:1255–1263
Dolja V, Meng B, Martelli G (2017) Evolutionary aspects of grapevine virology. Grapevine viruses: molecular biology, diagnostics and management. Springer, Cham, pp 659–688
Nakaune R, Toda S, Mochizuki M, Nakano M (2008) Identification and characterization of a new vitivirus from grapevine. Arch Virol 153:1827–1832
Al Rwahnih M, Sudarshana MR, Uyemoto JK, Rowhani A (2012) Complete genome sequence of a novel Vitivirus isolated from grapevine. J Virol 86:9545
Blouin AG, Chavan RR, Pearson MN, Macdiarmid RM, Cohen D (2012) Detection and characterisation of two novel vitiviruses infecting Actinidia. Arch Virol 157:713–722
Tzanetakis IE, Postman JD, Martin RR (2007) Identification, detection and transmission of a new vitivirus from Mentha. Arch Virol 152:2027–2033
Oliveira LM, Orilio AF, Inoue-Nagata AK, Nagata T, Blawid R (2017) A novel vitivirus-like sequence found in Arracacia xanthorrhiza plants by high throughput sequencing. Arch Virol 162:2141–2144
Candresse T, Theil S, Faure C, Marais A (2018) Determination of the complete genomic sequence of grapevine virus H, a novel vitivirus infecting grapevine. Arch Virol 163:277–280
Acknowledgements
This work was funded and supported by the Rod Bonfiglioli Scholarship from New Zealand Winegrowers Inc., and The New Zealand Institute for Plant & Food Research Limited Wine Research programme, funded by the Ministry for Business, Innovation and Employment Strategic Science Investment Fund. The Plant Biosecurity Cooperative Research Centre funded the small RNA sequencing and analysis (PBCRC2064). We would like to thank our Plant & Food Research colleagues: John Fletcher for the collection of samples, Sandi Keenan for her assistance in the sRNA extraction, and Kieren Arthur, and Dan Cohen as well as Mahmoud Khalifa (Damietta University, Egypt) for the useful comments on the manuscripts.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handeling Editor: Massimo Turina.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Blouin, A.G., Chooi, K.M., Warren, B. et al. Grapevine virus I, a putative new vitivirus detected in co-infection with grapevine virus G in New Zealand. Arch Virol 163, 1371–1374 (2018). https://doi.org/10.1007/s00705-018-3738-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3738-5