Abstract
Two double-stranded RNA (dsRNA) mycoviruses were found in isolate QSP5 of the rice blast fungus Magnaporthe oryzae. Sequence analysis of the two dsRNA mycoviruses revealed that one is closely related to Magnaporthe oryzae virus 2 (MoV2), and the other one is related to Magnaporthe oryzae chrysovirus 1-A (MoCV1-A). Therefore, they were named Magnaporthe oryzae virus 3 (MoV3) and Magnaporthe oryzae chrysovirus 1-C (MoCV1-C), respectively. In this paper, the molecular and structural characteristics of MoV3 were analyzed in detail. The full genome sequence (5181 bp) of MoV3 was obtained by cDNA cloning. Sequence analysis indicated that MoV3 has two overlapping open reading frames (ORF1 and ORF2). The 5′-proximal ORF1 encodes a putative coat protein (CP) with a molecular weight of 80,939 Da; the 3′-proximal ORF2 encodes a putative RNA-dependent RNA polymerase (RdRp) with a molecular weight of 90,506 Da. The stop codon of ORF1 overlaps the start codon of ORF2, with the tetranucleotide sequence AUGA, which is characteristic of members of the genus Victorivirus of the family Totiviridae. Phylogenetic analysis of RdRp and CP further supported the view that MoV3, a novel mycovirus, belongs to the genus Victorivirus of the family Totiviridae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Magnaporthe oryzae, a filamentous heterothallic ascomycete, is the most destructive pathogen of rice worldwide and the principal model organism for determining the molecular basis of fungal disease of plants. M. oryzae depends on its asexual spores (conidia) for disease establishment in nature. The conidia colonize leaves and produce necrotic lesions, or attack other parts of the plant at any stage of growth [10, 15]. Mycoviruses have been reported in many fungi [1, 4, 5]. Most mycoviruses found in filamentous fungi have double-stranded RNA (dsRNA) genomes; however, some novel single-stranded RNA (ssRNA) and single-stranded DNA (ssDNA) mycoviruses also have been reported [2, 18]. Four distinct viruses in the family Totiviridae and Chrysoviridae have been found in M. oryzae: Magnaporthe oryzae virus 1 and 2 (MoV1 and MoV2), belonging to the family Totiviridae, and Magnaporthe oryzae chrysovirus 1-A and 1-B (MoCV1-B and MoCV1-A), belonging to the family Chrysoviridae. The complete sequences of these viruses have been determined [8, 12–14, 17]. In this study, we report a novel mycovirus from strain QSP5 that was isolated from Hubei province in China. Sequence comparison and phylogenetic analysis revealed that MoV2 is closely related to members of the genus Victorivirus, which mainly infect filamentous fungi.
Materials and methods
The M. oryzae strain QSP5 was isolated from a lesion of a rice leaf collected in Hubei province, China, in 2011. The strain was stored on filter paper at −20 °C and cultured on potato dextrose agar (PDA) at 28 °C. dsRNA was extracted from the mycelia of strain QSP5 using cellulose (Sigma-Aldrich, Dorset, England) column chromatography [6, 7] and was detected using electrophoresis on 1.0 % agarose gels. Two dsRNA elements present in strain QSP5 were designated as Magnaporthe oryzae virus 3 (MoV3) and Magnaporthe oryzae chrysovirus 1-C (MoCV1-C) (Fig. 1a). To obtain the full dsRNA sequence of MoV3 in strain QSP5, the following methods were used: first, the purified dsRNA was used as a template for reverse transcription PCR (RT-PCR) amplification using a cDNA Synthesis Kit (Fermentas) with tagged random primer dN6 (5′-CGATCGATCATGATGCAATGCNNNNNN-3′) and a single specific primer (5′-CGATCGATCATGATGCAATGC-3′), which was based on the tagged random primer-dN6. The amplified PCR products were sequenced, analyzed and assembled using DNAMAN version 5.2.9 and the BLASTP program on the NCBI website [9, 16]. Second, regions corresponding to gaps in the sequences were obtained by RT-PCR using specific primers. Third, to obtain the 5′- and 3′-terminal sequence of dsRNA, an adaptor, pC3-T7loop (P-GGATCCCGGGAATTCGGTAATACGACTCACTATATTTTTATAGTGAGTCGTATTA-OH), was added to the terminal sequence of the purified dsRNA segments. The dsRNA with pC3-T7loop was then used as a template for RT-PCR using the specific primer pC2 (5′-CCGAATTCCCGGGATCC-3′), which was based on pC3-T7loop. Sequence analysis, alignments, phylogenetic analysis and identification of protein motifs was done by using the DNAMAN, CLUSTALX, MEGA5.0 and BLAST programs at the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [3, 9].
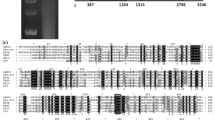
(a) dsRNAs in M. oryzae strain QSP5. Total dsRNA extracted from mycelia of strain QSP5. dsRNA samples were fractionated on a 1.0 % agarose gel and stained with ethidium bromide. The positions of host genomic DNA and viral dsRNAs (MoV3, MoCV1-C) of strain QSP5 are indicated. Lane M, Hind III-digested λ DNA marker. (b) Diagrammatic representation of the genomic organization of MoV3. (c) Amino acid sequence alignment of RdRps between MoV3 and selected viruses (GaRV-L1, MoV2, CmRV, HvV190S and MoV1) of the genus Victorivirus. Conserved motifs are motif I to VIII, asterisks represent identical residues, and colons represent similar residues
Sequence data
The full-length cDNA sequence of MoV3 has been deposited in the GenBank databases with accession number KP893140. The complete genome of MoV3 in strain QSP5 was 5181 bp, with a GC content of 61 %. Sequence analysis indicated that MoV3 has two overlapping open reading frames (ORF1 and ORF2). The 5′-proximal ORF1 (nt 283-2634) encodes a putative coat protein (CP) of 783 amino acid residues with a molecular weight of 80,939 Da that shows high similarity to the CPs of viruses in the family Totiviridae, particularly to those of Gremmeniella abietina RNA virus L1 (GaRV-L1, 73 % identity) [11], Magnaporthe oryzae virus 2 (MoV2, 67 % identity) [8], and Coniothyrium minitans RNA virus (CmRV, 63 % identity) [3]. Moreover,there is an Ala/Gly/Pro (A/G/P)-rich region near the C-terminal sequence (amino acid position from Gly711 to Gln783) of MoV3 CP, which is an unique molecular feature of members of the genus Victorivirus in the family Totiviridae [19]. The 3′-proximal ORF2 (nt 2631-5108) encodes a putative RNA-dependent RNA polymerase (RdRp) of 824 amino acid residues with a molecular weight of 90,506 Da, which showed a high similarity to the RdRps of viruses in the family Totiviridae, such as GaRV-L1 (58 % identity), MoV2 (52 % identity) and CmRV (49 % identity). According to the ICTV rules for species demarcation in the genus Victorivirus, sequence identity less than 60 % in either the CP or the RdRP protein indicates that the viruses belong to different species [19]. Although the CP sequence of MoV3 shares 67 % identity with MoV2, the RdRp sequence shares only 52 % identity with MoV2, which is less than the 60 % cutoff for species delimitation. Thus, MoV2 could be a new species in the genus Victorivirus. The stop codon of ORF1 overlaps the start codon of ORF2 with the tetranucleotide sequence AUGA, which is characteristic of the genus Victorivirus of the family Totiviridae. The 5′ untranslated region (UTR) and 3′-UTR contains 282 and 73 nucleotides, respectively (Fig. 1b).
In addition, a multiple sequence alignment of the deduced amino acid sequences of the RdRps of MoV3 and five other victoriviruses (GaRV-L1, MoV2, CmRV, HvV190S and MoV1) in family Totiviridae revealed that MoV3 has eight conserved motifs that are typical of members of the family Totiviridae. In addition, the eight conserved regions exhibited high sequence similarity among these viruses (Fig. 1c).
Phylogenetic analysis of the RdRps between MoV3 and selected viruses in the families Totiviridae and Chrysoviridae showed that the putative RdRp of MoV3 clusters closely with the members of genus Victorivirus (GaRV-L1, MoV2, CmRV, HvV190S and MoV1). Furthermore, phylogenetic analysis of the CP of MoV3 and that of viruses in the families Totiviridae and Partitiviridae also indicated that the putative CP of MoV3 clusters with members of genus Victorivirus (Fig. 2).
Phylogenetic analysis of the RdRp and CP genes of MoV3 and related viruses in the families Totiviridae, Chrysoviridae and Partitiviridae. The unrooted phylogenetic tree was constructed by the neighbor-joining method using the program MEGA 5.0. Bootstrap values (1000 replicates) are shown at the nodes, and the scale bar (0.1) corresponds to genetic distance. The position of MoV3 is indicated by a red star. The sequences of the RdRp and CP were obtained from the GenBank database. The mycoviruses are as follows (abbreviation; GenBank accession no.): Gremmeniella abietina RNA virus L1 (GaRV-L1; AAK11656.1), Coniothyrium minitans RNA virus (MoV2; YP_001649206.1), Coniothyrium minitans RNA virus (CmRV; YP_392467.1), Helminthosporium victoriae virus 190S (HvV190S; NP_619670.2), Magnaporthe oryzae virus 1 (MoV1; P_122352.1), Leishmania RNA virus 1-1 (LRV1-1; NP_041191.1), Leishmania RNA virus 2-1 (LRV2-1; NP_043465.1), Trichomonas vaginalis virus 1 (TvV1; ABF57713.1), Trichomonas vaginalis virus 2 (TvV2; AED99806.1), Trichomonas vaginalis virus 4 (TvV4; AED99794.1), Trichomonas vaginalis virus 3 (TvV3; AED99800.1), Saccharomyces cerevisiae virus L-A (ScV-L-A; NP_620495.1), Saccharomyces cerevisiae virus L-BC (ScV-L-BC; NP_042581.1), Cryphonectria nitschkei chrysovirus 1 (CnCV1; ACT79256.1), Verticillium dahliae chrysovirus 1 (VdCV1; ADG21213.1)
Our present results showed that MoV3 is a new member of the genus Victorivirus of the family Totiviridae. Our report provides preliminary evidence that two distinct viruses, MoV3 and MoCV1-C, from the families Totiviridae and Chrysoviridae, could exist in the same host, which is similar to what has been reported previously for Helminthosporium victoriae, which can be co-infected by the victorivirus Helminthosporium victoriae virus 190S and the chrysovirus Helminthosporium victoriae virus 145S [5].
References
Bao X, Roossinck MJ (2013) Multiplexed interactions: viruses of endophytic fungi. Adv Virus Res (San Diego, CA, Elsevier) 86(86):37–57
Buck KW (1986) Fungal virology an overview. In: Buck KW (ed) Fungal virology. CRC Press, Boca Raton, pp 1–84
Cheng J, Jiang D, Fu Y, Li G, Peng Y, Ghabrial SA (2003) Molecular characterization of a dsRNA totivirus infecting the sclerotial parasite Coniothyrium minitans. Virus Res 93:41–50
Dawe AL, Nuss DL (2013) Hypovirus molecular biology: from Koch’s postulates to host self-recognition genes that restrict virus transmission. Adv Virus Res (San Diego, CA, Elsevier) 86(86):109–147
Ghabrial SA, Dunn SE, Li H, Xie J, Baker TS (2013) Viruses of Helminthosporium (Cochlioblus) victoriae. Adv Virus Res (San Diego, CA, Elsevier) 86(86):289–325
Ghabrial SA, Suzuki N (2010) Fungal viruses. In: Van Mahy BWJ, Regenmortel MHV (eds) Desk encyclopedia of plant and fungal virology. Elsevier, Oxford, pp 517–524
Liu H, Fu Y, Jiang D, Li G, Xie J, Peng Y, Yi X, Ghabrial SA (2009) A novel mycovirus that is related to the human pathogen Hepatitis E virus and rubi-like viruses. J Virol 83(4):1981–1991
Maejima K, Himeno M, Komatsu K, Kakizawa S, Yamaji Y, Hamamoto H, Namba S (2008) Complete nucleotide sequence of a new double-stranded RNA virus from the rice blast fungus, Magnaporthe oryzae. Arch Virol 153:389–391
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Talbot NJ (2003) On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu Rev Microbiol 57:177–202
Tuomivirta TT, Hantula J (2003) Two unrelated double-stranded RNA molecule patterns in Gremmeniella abietina type A code for putative viruses of the families Totiviridae and Partitiviridae. Arch Virol 148:2293–2305
Urayama S, Kato S, Suzuki Y, Aoki N, Tuong LM, Arie T, Teraoka T, Fukuhara T, Moriyama H (2010) Mycoviruses related to chrysovirus affect vegetative growth in the rice blast fungus Magnaporthe oryzae. J Gen Virol 91:3085–3094
Urayama S, Ohta T, Onozuka N, Sakoda H, Fukuhara T, Arie T, Teraoka T, Moriyama H (2012) Characterization of Magnaporthe oryzae chrysovirus 1 structural proteins and their expression in Saccharomyces cerevisiae. J Virol 86(15):8287–8295
Urayama S, Sakoda H, Takai R, Katoh Y, Minh LT, Fukuhara T, Arie T, Teraoka T, Moriyama H (2014) A dsRNA mycovirus, Magnaporthe oryzae chrysovirus1-B, suppresses vegetative growth and development of the rice blast fungus. Virology 448:265–273
Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus Magnaporthe oryzae. Annu Rev Microbiol 29:443–467
Xie J, Xiao X, Fu Y, Liu H, Cheng J, Ghabrial SA, Li G, Jiang D (2011) A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology 418:49–56
Yokoi T, Yamashita S, Hibi T (2007) The nucleotide sequence and genome organization of Magnaporthe oryzae virus 1. Arch Virol 152:2265–2269
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X (2010) A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA 107:8387–8392
Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC (2011) Totiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, pp 639–650
Acknowledgments
This work was supported by the Commonweal Specialized Research Fund of China Agriculture (201203014), the National Natural Science Foundation (31201478), and the China Postdoctoral Science Foundation (2012M520782).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, L., Hu, Y., Liu, L. et al. Genomic organization of a novel victorivirus from the rice blast fungus Magnaporthe oryzae . Arch Virol 160, 2907–2910 (2015). https://doi.org/10.1007/s00705-015-2562-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-015-2562-4