Abstract
Nonstructural proteins of members of the family Reoviridae are believed to play significant roles in the virus replication cycle. Phylogenetic analyses indicate that the nonstructural protein NS80 of grass carp reovirus, encoded by a gene of Segment 4 (S4), is a primary determinant that is related to the formation of viroplasmic inclusion bodies (VIB), where viral replication and assembly are thought to occur. To understand the role of the NS80 protein in viral replication, an initial investigation of NS80 gene expression in both infected and transfected cells was conducted. Transmission electron microscopy results indicate that replication and assembly of GCRV occur within VIB-like structures in the perinuclear region of the cell cytoplasm. Furthermore, expression of the S4 gene in infected cells was detected with an NS80-specific antibody by western blot and immunofluorescence. Moreover, globular VIB-like structures were observed when expressing GFP-derived full-length NS80 (pEGFP-C1/NS80) and recombinants containing the C-terminal conserved region (pEGFP-C1/NS80335–724) in transfected Vero. No such structures were detected in cells transfected with an N-terminal recombinant (pEGFP-C1/NS801–334), suggesting that the NS80 C-terminal conserved region may be involved in the formation of inclusion structures. These data provide a foundation for further functional studies of NS80 related to viral inclusion formation in viral replication.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
All members of the family Reoviridae possess a segmented double-stranded RNA (dsRNA) genome encased in multiple concentric protein capsids. They are non-enveloped, icosahedral virions with a double capsid structure, consisting of a genome comprised of 9–12 segments of dsRNA in the core. During replication and assembly of these viruses, viroplasmic inclusion bodies (VIB), or viral factories (VF), are formed throughout the cytoplasm of infected cells [7, 8, 11, 12, 32], suggesting that VIB-like structures are essential for the viral replication cycle. These VIB-like structures have a peculiarly dense consistency that distinguishes them from the adjacent cytoplasm and causes them to appear highly refractile when viewed by phase-contrast microscopy, and they contain fully or partially assembled viral particles, viral proteins, dsRNA and microtubules [4–7, 20, 23, 26]. Recent studies on mammalian orthoreoviruses (MRV) and avian orthoreoviruses (ARV) as well as other genera in the family Reoviridae, such as rotaviruses and orbiviruses [8, 12], have shown that the orthoreovirus nonstructural protein μNS plays a major role in the formation of cytoplasmic inclusion structures.
The family Reoviridae currently includes 15 recognized genera of viruses [3, 22], including the genus Aquareovirus. Six aquareovirus species have been classified by the International Committee on Taxonomy of Viruses (ICTV), from Aquareovirus A (AQRV-A) to Aquareovirus F (AQRV-F) [3, 21], and an additional, seventh species, named Aquareovirus G, has also been recognized recently [24]. Like turreted MRV and other members of the family Reoviridae, GCRV, which forms a multilayered spherically structured particle, has a genome of 11 dsRNA segments, which encode 7 structural proteins (VP1–VP7) and 5 nonstructural proteins. The mature viral particle consists of five core proteins and two outer capsid proteins, while the five nonstructural proteins (NS80, NS38, NS31, NS26, NS16) may play roles in the viral replication cycle [2]. Unlike the nonfusogenic MRV, GCRV can produce large syncytia as a typical cytopathic effect (CPE) in Ctenopharyngodon idellus kidney (CIK) cell lines during infection, and mature virions are efficiently released from infected cells by lysis [14]. Studies on GCRV and other aquareovirus genomes have revealed that there is a close evolutionary relationship between members of the genera Orthoreovirus and Aquareovirus [2, 15]. Recent progress on structural reconstruction of single virus particles has further confirmed this high level of identity at the protein level [9, 10, 17], especially for the core proteins, including the functional and enzymatic domains, which are involved in maintaining the stability of the inner core shell and endogenous transcription activity among dsRNA viruses in general. Despite the significant divergence that exists between GCRV and MRV in the outer capsid protein components, the functional domains related to cell entry remain conserved. In addition to the structural proteins, non-structural proteins are believed to be indispensable for viral replication and assembly of the nascent particle. For example, NS80, which has been shown to play a key role in the formation of factory-like inclusion structures, is critical for the viral replication cycle.
Substantial progress has been made in clarifying nonstructural protein function in viral replication for members of several genera in the family Reoviridae, including Orthoreovirus, Rotavirus, Orbivirus and Phytoreovirus [1, 4–8, 11, 18, 20, 22, 23, 29]. In MRV and ARV, the nonstructural protein μNS expressed alone in transfected cells forms VIB-like structures that, when viewed by phase-contrast microscopy, appear to be similar to VF formed in infected cells [25], suggesting that μNS is able to form the matrix of the factories. As a step toward understanding the molecular basis of viral factory formation in viral replication and assembly, an investigation of NS80 gene expression in both infected cells and transfected cells was carried out in this study. Ultrathin-section images obtained by transmission electron microscopy (TEM) showed that replication and assembly of GCRV took place within VIB-like structures in the perinuclear region of the cytoplasm. Additionally, expression of the NS80 gene in both infected and transfected cells was detected with specific polyclonal NS80 antibodies by both western blot (WB) and immunofluorescence (IF). Furthermore, inclusion-like structures were observed in Vero cells transfected with recombinant plasmids encoding GFP-fused, full-length NS80 (pEGFP-C1/NS80) and the conserved carboxyl-terminal region (pEGFP-C1/NS80335–724). The results provide a foundation for further functional analysis of the NS80 protein.
Materials and methods
Cells and virus
A CIK cell line was prepared for the GCRV infection assays in this study. The cells were grown at 28°C in monolayers in Eagle’s minimum essential medium (MEM, Invitrogen) containing 2 mM l-glutamine, supplemented with 10% of fetal bovine serum (MEM-10). Vero cells used for the transfection assay were grown at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, USA) supplemented with 10% fetal bovine serum, and 2 mM l-glutamine. Both penicillin (100 U/ml) and streptomycin (100 μg/ml) antibiotics were added to the MEM and DMEM medium during cell cultivation. GCRV-873, the original isolate of GCRV, grown in CIK monolayer cells as described previously [14, 19], was used in this study.
Construction of recombinant plasmids
To construct a recombinant plasmid containing the NS80 gene, RT-PCR was performed by using the purified whole GCRV genome as a template to amplify the NS80 gene encoded on GCRV Segment 4 (S4). The primers targeting GCRV S4 were designed based on GenBank sequence AF403390 and contained specific restriction enzyme digestion sites. The recombinant plasmid expressing the C-terminal region of NS80 in E. coli was named pR/NS80335–742 and has been described previously [13]. To express the whole NS80 protein, the full-length S4 gene was cloned into the pRSET-A vector (Invitrogen, Carlsbad, USA) in this study, and this clone was named pR/NS80. The primer pair used for amplification of the full-length S4 gene was as follows: sense primer 5′CATGGATCCAAGAGCTTTGGAAGTAAC3′ (BamHI site underlined), and anti-sense primer 5′GCTGAATTCGACACAGAAACACAGAGC3′ (EcoRI site underlined). The remaining cloning steps are the same as reported previously [13]. Recombinant plasmids that contained the gene of interest were sequenced by Invitrogen Biotechnology Inc. (Shanghai, China).
To generate constructs for expression of green fluorescence protein (GFP) fused to the N terminus, three primer pairs (GCRV-S4-SN1/AS2, GCRV-S4-SN1/AS1, GCRV-S4-SN2/AS2), including forward primer 1 (GCRV-S4-SN1) 5′C ATGAATTCGAAGAGCTTTGGAAGTAAC3′ (EcoRI site underlined), forward primer 2 (GCRV-S4-SN2) 5′C GAGAATTCTCCCTCCTTACCCTTC3′ (EcoRI site underlined), reverse primer1 (GCRV-S4-AS1):5′CACGGATCCGATGTCTTCGGTGGGAGC3′ (BamHI site underlined), and reverse primer 2 (GCRV-S4-AS2) 5′CAAGGATCCGACACAGAAACACAGAGC3′ (BamHI site underlined), were designed for RT-PCR amplification to generate full-length S4, an S4 N-terminal fragment, and an S4 C-terminal fragment, respectively. The amplified fragments were cloned into a pEGFP-C1 vector (Clontech) that had been cut with EcoRI and BamHI enzymes to generate pEGFP-C1/NS80, pEGFP-C1/NS801–334 and pEGFP-C1/NS80335–742 recombinants. E. coli DH5α cells were then transformed with the ligated products for further identification. The correctness of each construct was confirmed by sequencing (Invitrogen Biotechnology Inc., Shanghai, China).
Expression of recombinant NS80 in E. coli and preparation of antibody
To express recombinant NS80 in E. coli, BL21(DE3) cells were transformed with the pR/NS80 construct, and in vitro expression and purification of NS80 were conducted as described previously [13]. Antibodies against the recombinant NS80 protein were prepared by injecting New Zealand white rabbits subcutaneously with a mixed emulsion of 200 μl of purified recombinant NS80 solution (about 0.5 μg/μl) and an equal volume of Freund’s complete aduvant (FCA), followed by two intramuscular booster injections with an equal volume every 2 weeks. Three days later, after a final booster injection, the rabbits were bled, and sera were separated, aliquoted and stored at −80°C for further use. The antibody titer was determined by ELISA [33].
Virus replication and transmission electron microscopy examination
Virus infection of CIK cells was conducted as described elsewhere [14, 19]. When a CPE was observed in virus-infected cells, the culture supernatant of the lysed cells was harvested. The collected GCRV culture suspension was then stored at −30°C or at 4°C prior to use. For sequential analysis of GCRV replication in CIK cells, infected monolayer cells were collected at different intervals postinfection (p.i.), collected by centrifugation at 12,000 rpm for 10 min, and then stored at −30°C for further immunoblotting analysis. For ultrathin section sample preparation, harvested cells were fixed with 2.5% glutaraldehyde in 0.2 M sodium cacodylate buffer (pH 7.4) for 24 h at 4°C. Mock-infected CIK cell monolayers were used as a control.
GCRV particles harvested from infected culture supernatant were purified by either sucrose gradient centrifugation or CsCl density gradient centrifugation as described elsewhere [16, 17]. For TEM examination of purified virus, virion samples were deposited onto formvar-coated, carbon-stabilized copper grids (200-mesh) before they were negatively stained with 3% (w/v) phosphotungstic acid (PTA, pH 6.8). For ultrathin section samples, GCRV-infected CIK cells pre-fixed with 2.5% paraformaldehyde were sliced, mounted on 200-mesh 0.25% formvar-coated copper grids, and double stained with 2% uranyl acetate and lead citrate. All sample grids were examined using a transmission electron microscope (Hitachi 7000-FA).
Infection and IF microscopy
To detect in vivo-expressed NS80 protein by IF, CIK cells were seeded on the day before infection at a density of 1.0 × 104 cells/cm2 in six-well plates (9.6 cm2 per well) containing glass coverslips. GCRV873, a third-passage infected CIK cell lysate stock of plaque-purified suspension, was used as the virus strain [14]. A virus stock at an MOI of 10–20 PFU/cell was inoculated onto confluent monolayers of CIK. After about 30–60 min of viral adsorption, cells were then washed with phosphate-buffered saline (PBS, 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1 mM KH2PO4, pH 7.5) and further incubated at 28°C for maintenance in Eagle’s MEM with 2% of fetal bovine serum (MEM-2) before being processed for subsequent experiments. The infected monolayer cells were collected at 6-h intervals beginning from 0 to 24 h p.i. and fixed for 10 min at −20°C in 100% methanol and then permeabilized, blocked, incubated with antibodies, and mounted as described previously [26]. Primary NS80 specific antibody was diluted in PBST and incubated with cells for 30–60 min at room temperature. After washing three times with PBS, secondary fluorescein isothiocyanate (FITC)-tagged antibodies diluted in PBST were added and incubated with cells for 30 min at room temperature. Samples were examined using an Olympus-IX51 inverted microscope equipped with phase and fluorescence optics, and images were collected digitally. All of the images were processed and prepared for presentation by using Image Pro-Eexpress 6.3 (Olympus) and Photoshop (Adobe Systems).
Transient expression of recombinant NS80 proteins
After confirming the correct orientation of the NS80 gene in the pEGFP vector by both restriction analysis and nucleotide sequencing, the pEGFP-C1/NS80, pEGFP-C1/NS801–334 and pEGFP-C1/NS80335–742 recombinants were purified using a QIAfilter Plasmid Maxi Kit (QIAGEN) and used to transfect Vero cells according to the transfection kit user manual (Lipofectamine 2000, Invitrogen). Briefly, 1 day before transfection, 0.5 × 105 to 2 × 105 cells were cultured in six-well plates growing in DMEM medium without antibiotics so that cells were 80–85% confluent at the time of transfection. About 4.0 μg of plasmid DNA per well was diluted in 250 μl DMEM and then mixed with 10 μl of Lipofectamine per well, diluted with the same medium. After a 20-min incubation, the mixtures were added to each well containing cells and medium. Cells were incubated at 37°C, and the medium was changed at 6 h post-transfection (pt). Transfected cells were observed around 18–24 h pt. GFP fusion proteins were detected by their intrinsic GFP fluorescence. Cell images were visualized using an Olympus-IX51 inverted microscope. In addition, the transfected cells were collected and lysed for further immunoblotting as described elsewhere [31].
SDS-PAGE and immunoblotting analysis
To analyze viral proteins expressed in infected cells, the collected cell lysates were pelleted. The pelleted cells were resuspended in 60 μl of 1× PBS, lysed in sample buffer, boiled for 2 min, and subjected to SDS-PAGE. All protein samples were detected by staining with Coomassie brilliant blue R-250 (Sigma) or were transferred to a polyvinylidene fluoride (PVDF) transfer membrane using a semi-dry transfer cell (Bio-Rad) and visualized by western blotting. NS80-specific antisera (1:500) or mouse monoclonal GFP antibody (1:1,000) were employed as primary antibodies. An alkaline-phosphatase-coupled goat anti-rabbit IgG (Sigma) or rabbit anti-mouse was used as the secondary antibody at a dilution of 1:2,000. The result was observed by developing with NBT/BCIP AP substrate solution (Promega) according to the manufacturer’s instructions.
Sequence comparisons
To determine conserved domains, GCRV NS80 (accession no. AF403390) was compared with the following homologous sequences: NS1 of chum salmon reovirus (aquareovirus C, CSRV, accession no. NC_007585), and μNS of mammalian orthoreoviruse T1L (accession no. AF174382), μNS of avian orthoreovirus S1133 (accession no. AY608700). Data were analyzed by Protein Blast in NCBI (http://blast.ncbi.nlm.nih.gov).
Results
Viroplasmic inclusions observed during virus replication
To examine GCRV morphogenesis during replication, ultrathin-section images of virus-infected cells were viewed by TEM. As shown in Fig. 1, during GCRV replication, virus particles were found in interior regions called viroplasmic inclusions, which are not bound by a cellular membrane but are distinct from the surrounding cytoplasm. Early virus particles (about 4–8 h p.i.) of different sizes (about 35–55 nm in diameter) were observed within the dense viroplasms, as shown in Fig. 1a and b. As the infection progressed, the size and number of viroplasmic inclusions increased (Fig. 1c). Generally, inclusions were typically concentrated in the perinuclear region of the cytoplasm. At the time of late infection (18 h), mature virions were formed with an overall size of about 70–80 nm (Fig. 1d). Eventually, progeny virions can lead to the breakdown of infected cells so that mature particles are released efficiently into the cell culture supernatant to initiate the next cycle of replication. It appeared that once assembly had been achieved, the dense viroplasmic inclusions become thin and the mature virion was formed, indicating that virus replication and assembly occurred within the viroplasm-like inclusions (Fig. 1d).
Expression of full-length NS80 in E. coli
Protein NS80, which is encoded by the GCRV S4 gene, consists of 742 amino acids (about 80 kDa). When BLAST analysis was used to compare the NS80 analogue with that of CSRV in the genus Aquareovirus and MRV and ARV in the genus Orthoreovirus, identities could be found mostly within the C-terminal region of the protein (Fig. 2a). Specially, the proteins share two coiled-coil regions (513–548 and 615–700AA) in their carboxyl-proximal region, which have been shown to be important in inclusion formation by MRV [7]. Based on this analysis, to generate specific antiserum of NS80, the C-terminal region of NS80 recombinant (pR/NS80335–742) was first constructed as described previously [13].
Alignment of GCRV NS80 with analogous proteins and expression of NS80 protein. a Alignment of GCRV NS80 with analogous proteins of CSRV, ARV and MRV (see “Materials and methods” for accession numbers). The alignment shows that the C-terminal region of NS80 has a high alignment score with CSRV, ARV and MRV, and this region is conserved among the different viruses. b SDS-PAGE of GCRV NS80 expressed in E. coli. M standard protein marker; Lanes 1 and 2 expressed NS80 protein cell lysate pellet and supernatant induced 3 h by IPTG, respectively. Lanes 3 and 4 expressed NS80335–742 protein cell lysate pellet and supernatant induced 3 h by IPTG, respectively. Lane 5 cell lysate pellet of expressed empty vector (pRSET-A) induced 3 h by IPTG. c Western blot assay using NS80335–742 polyclonal antibody. M pre-stained standard protein marker. 1 cell lysate of expressed empty vector. 2 and 3 expressed NS80 cell lysate and purified NS80. 4 mock-infected CIK cell, 5 GCRV-infected CIK cell lysate at 24 h p.i.
In this study, we constructed a full-length NS80 recombinant, named pR/NS80. As shown in Fig. 2b, the full-length NS80 recombinant was successfully expressed in E. coli, which produced a major protein of approximately 83 kDa (including an N-terminal tag of about 3 kDa) that was not present in the empty vector cell lysate, but the expression level was somewhat lower than that of NS80335–742. The identity of this protein was further confirmed by immunoassays using a his-tag antibody and previously prepared NS80335–742 polyclonal antiserum [13]. Immunoblotting showed that the expressed NS80 cell lysate and purified NS80 protein (SDS-PAGE data not shown) were able to bind to an NS80335–742-specific rabbit antibody (Fig. 2c), indicating correct expression of recombinant NS80 protein. In addition, the product of the S4 gene expressed in GCRV infected cells cross-reacted with the NS80335–742 antibody, suggesting that expression of the NS80 gene could be detected in GCRV-infected cells by using the prepared specific NS80335–742 antibody. At the same time, a small protein (about 70 kDa) was also detected with the specific antibody. To evaluate the titers of the prepared antibodies, both NS80 and NS80335–742 antisera were examined by ELISA (data not shown). Both of the antibodies were used in subsequent immunoassays.
NS80 expression detected in GCRV-infected cells
As reported previously, GCRV can produce large syncytia as a typical CPE when a sensitive cell line is infected [14], in contrast to nonfosogenic MRV. To investigate whether GCRV replication is related to NS80 gene expression in infected cells, both immunoblotting and indirect IF were conducted (Figs. 3, 4). SDS-PAGE and western blot analysis (Fig. 3a, b) showed that expression of the S4 gene can be detected, resulting in a protein with a molecular weight around 80 kDa at 6,12,18 h p.i., suggesting that it is the full-length NS80. However, a shorter polypeptide (about 70 kDa) was also detected in all of the infected cell lysate samples (Fig. 2c, lane 5, Fig. 3b, lanes 2–5), which might be truncated NS80 or a degraded form of full-length NS80 (see “Discussion”). Furthermore, high-level expression of NS80 with inclusion-like structures was observed at 12 and 18 h p.i. by phase-contrast and IF microscopy (Fig. 4). NS80 could also be detected at 24 h p.i. in infected cell lysates by western blot (Fig. 2c, lane 5), but it was not clearly detected by IF at 24 h p.i. because most of the nascent virions had been released from cells at this point. No NS80 protein could be detected in mock-infected cells or purified virus samples (Fig. 3, lanes 1, and 6).
SDS-PAGE and immunoblotting analysis of NS80 expression in infected cells. a SDS-PAGE analysis of NS80 expression in infected cells at 0, 6, 12, and 18 h p.i. M standard protein marker; 1–4 GCRV-infected CIK cells at 0, 6, 12, and 18 h p.i. 5 Expressed NS80 in E. coli cell lysate (arrowhead). 6 Purified GCRV virion. b NS80 protein was detected in infected cells by immunoblotting analysis with protein samples corresponding to those in a with NS80335–742 antibody, Lanes 1–6 in b correspond to lanes 1–6 in a
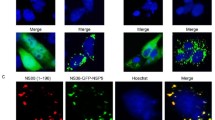
Immunofluorescence analysis of NS80 expression in infected cells. CIK cells were infected with GCRV at a multiplicity of infection of 10 PFU/cell. Infected cells were fixed at 12 and 18 h p.i. and immunostained with rabbit anti-NS80 serum followed by goat anti-rabbit IgG conjugated to FITC. Mock-infected cells are shown in the upper panels, and cells infected with virus are shown in the middle and bottom panels. Left, middle columns are phase and fluorescence images, respectively. Merged images are shown in the right column. Arrows indicate inclusion-like structures. Scale bars 10 μm
Inclusions formed in NS80-transfected cells
By analogy to μNS of MRV, GCRV NS80 is thought to be a key protein in the formation of viral inclusions. To test whether the NS80 protein alone (i.e., in the absence of any other viral components in transfected cells) is able to form inclusions, Vero cells were lipo-transfected with three GFP-tagged recombinants, pEGFP-C1/NS80, pEGFP-C1/NS801–334 and pEGFP-C1/NS80335–742. As predicted, the recombinant full-length GFP/NS80 and GFP/NS80335–742 C-terminal proteins were able to form factory-like inclusions in transfected cells. Globular inclusion-like structures were observed around 18–24 h pt due to their intrinsic green fluorescence (Fig. 5a), and these appeared similar to those formed by μNS in MRV-infected cells [7, 22], whereas no inclusion structures were detected in cells transfected with the GFP-tagged N-terminal conserved region recombinant (pEGFP-C1/NS801–334). Likewise, green fluorescence from transfected empty EGFP-C1 was distributed throughout the cytoplasm and nucleus. To determine the correctness of the expressed recombinant protein, all of the cell lysates were subjected to SDS-PAGE, followed by immunoblotting with both GFP-specific mouse MAb and rabbit anti-NS80 serum. The results verified that the expressed GFP-derived fusion proteins from each construct were not only the right size but also produced a serological reaction with NS80 polyclonal antibody (Fig. 5b, c), indicating that the protein was expressed efficiently. This result suggests that NS80 or the C-terminal region alone is able to form factories or inclusion-like structures in transfected cells.
Fluorescence microscopy and Immunoblotting of GFP-tagged NS80 derivatives. Vero cells were transfected with plasmids to express GFP-tagged NS80, NS801–334 and NS80335–742 fusion protein and then analyzed at 20 h p.t. by using fluorescence microscopy and western blotting. a Observation of expressed recombinant protein by fluorescence microscopy. A viral inclusion-like structure can be detected in GFP-NS80- and GFP-NS80335–742-transfected cells. As shown in the upper left and upper right panels, inclusions were visualized using the inherent fluorescence of EGFP. Vero cells transfected with GFP-NS801–334 and empty GFP vector are shown in the lower left and lower right panels, in which green fluorescence is dispersed throughout the cytoplasm and nucleus. b, c Immunoblotting assay using anti-GFP MAb and anti-NS80 polyclonal serum as the primary antibody and alkaline-phosphatase-coupled goat anti-rabbit IgG or rabbit anti-mouse as the secondary antibody. M pre-stained standard protein marker; 1 GFP-NS801–334, 2 mock-infected Vero cells, 3 GFP-NS80335–742, 4 GFP-NS80, 5 GFP. The arrows show expressed protein of interest. Bars 10 μm
Discussion
Usually, virus replication and assembly occur in specific locations within infected cells. For example, in herpes simplex virus (dsDNA genome; family Herpesviridae), this occurs in nuclear inclusions, and in flock house virus (ssRNA genome; family Nodaviridae), it occurs on the outer mitochondrial membranes [23, 28]. For viruses in the family Reoviridae, previous investigations have indicated that the cytoplasmic phase-dense inclusions in reovirus-infected cells are formed and located at perinuclear sites during viral replication and assembly [7, 23, 25]. In MRV and ARV, the nonstructural protein μNS has been identified to be the most important key factor in forming the matrix of these viral inclusions and recruiting other components to viroplasma-like structures [4–7, 20, 22, 30, 31]. However, the role of nonstructural proteins in aquareovirus virus replication has been poorly characterized to date.
GCRV, which is regarded as the most pathogenic of all of the aqureoviruses that have been isolated [27], is a tentative member of the family Reoviridae [2, 3, 21, 24]. Sequence analysis indicates that GCRV and MRV have a common evolutionary origin although they have different host species. Among the 12 proteins of GCRV that have been identified, 5 nonstructural proteins appear at different stages in the viral life cycle. Of these nonstructural proteins, NS80 has been identified as a homolog of the μNS protein in MRV. To identify specific residues in NS80 that may be required for inclusion formation, we compared the deduced protein sequences of NS80 homologs from mammalian and ARV. Although BLAST analysis indicated significant divergence in the N-terminal region of NS80, there were several conserved regions that were mainly concentrated in the C-terminal region. Other aquareoviruses, such as golden shiner reovirus (GSRV), belonging to Aquareovirus group C and CSRV, belonging to Aquareovirus group A, also share conserved C-terminal domains. A C-terminal Leu residue is also conserved among the μNS proteins of MRV and ARV [34]. These conserved regions in MRV have been identified to be critical for inclusion formation [7, 20, 22]. Based on these findings, in addition to the full-length NS80 protein, we designed a truncated NS80 recombinant from aa 335–742 to determine whether the C-terminal conserved region is essential for inclusion formation. Our results indicate that NS80335–742 is sufficient for the formation of inclusion bodies in transfected cells.
It is interesting to note that we detected an additional band by immunoblotting, with a size around 70 kDa in the GCRV-infected cell lysate, and based on its molecular weight, this might be a C-terminal form of the NS80 translation product that begins at an internal in-frame initiation codon in the S4 mRNA, or it might just be a degraded form of full-length NS80. A truncated form of μNS, μNSC, which exists in MRV and lacks about 5 kDa from its N terminus, could also be detected in infected cells [5]. The origin of μNSC is not certain, but it has been proposed that it is a consequence of either secondary initiation or cleavage near Met41 in μNS [5, 6]. A similar situation has also been found in ARV, with two μNS isoforms recognized by μNS-specific antibodies. In addition to the 70-kDa full-length μNS in ARV, a 10-kDa smaller protein (about 60 kDa) was also detected in ARV infected cells that was not found to associate with the ARV σNS protein, suggesting that the activities of μNS and μNSC may function at different stages of regulation of viral replication and assembly [31]. Similar to μNSC in MRV or ARV, the roles of truncated NS80 and the possible NS80C polypeptide detected in this study may warrant further investigation.
The experiments described in this report were aimed at providing insight into the function of the GCRV nonstructural protein NS80 in viral replication. It appears that the GCRV particles were formed in the interior region of viroplasm-like inclusions. NS80, which has been deduced to be a key protein in virus replication, was detected by immunoblotting and indirect IF in both infected and transfected cells using polyclonal NS80 antiserum. Moreover, as expected, globular-like inclusions dispersed throughout the cytoplasm were observed in cells transfected with GFP-tagged full-length NS80 and a C-terminal region recombinant, suggesting that the C-terminal region from aa 335 to 742 is sufficient to form inclusions. The data provided in this study will allow us to further test the function of NS80 in the formation of the viral factory matrix in viral replication and assembly.
References
Arnold MM, Murray KE, Nibert ML (2008) Formation of the factory matrix is an important, though not a sufficient function of nonstructural protein μNS during reovirus infection. Virology 375:412–423
Attoui H, Fang Q, Mohd JF, Cantaloube JF, Biagini P, Micco P, Lamballerie X (2002) Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J Gen Virol 83:1941–1951
Attoui H, Jaafar FM, Belhouchet M, Biagini P, Cantaloube JF, de Micco P, de Lamballerie X (2005) Expansion of family Reoviridae to include nine-segmented dsRNA viruses: isolation and characterization of a new virus designated Aedes pseudoscutellaris reovirus assigned to a proposed genus Dinovernavirus. Virology 343:212–223
Broering TJ, Mccutcheon AM, Centonze V, Nibert ML (2000) Reovirus nonstructural protein μNS binds to core particles but does not inhibit their transcription and capping activities. J Virol 74(12):5516–5524
Broering TJ, Parker JSL, Joyce PL, Kim J, Nibert ML (2002) Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J Virol 76:8285–8297
Broering TJ, Kim J, Miller CL, Piggott CDS, Dinoso JB, Nibert ML, Parker JSL (2004) Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J Virol 87(4):1882–1892
Broering TJ, Arnold MM, Miller CL, Hurt JA, Joyce PL, Nibert ML (2005) Carboxyl-proximal regions of reovirus nonstructural protein μNS necessary and sufficient for forming factory-like inclusions. J Virol 79:6194–6206
Brookes SM, Hyatt AD, Eaton BT (1993) Characterization of virus inclusion bodies in bluetongue virus-infected cells. J Gen Virol 74:525–530
Cheng LP, Fang Q, Shah S, Atanasov IC, Zhou ZH (2008) Subnanometer-resolution structures of the grass carp reovirus core and virion. J Mol Biol 382:213–222
Cheng LP, Zhu J, Hui WH, Zhang XK, Honig B, Fang Q, Zhou ZH (2010) Backbone model of an aquareovirus virion by cryo-electron microscopy and bioinformatics. J Mol Biol 397:852–863
Eichwald C, Rodriguez JF, Burrone OR (2004) Characterization of rotavirus NSP2/NSP5 interactions and the dynamics of viroplasm formation. J Gen Virol 85:625–634
Estes MK (2001) Rotaviruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (eds) Fields virology, 4th edn. Lippincott Williams & Wilkins, Philadelphia, pp 1747–1786
Fan C, Zhang LL, Lei CF, Fang Q (2009) Expression and identification of inclusion forming-related domain of NS80 nonstructural protein of grass carp reovirus. Virol Sin 24:194–201
Fang Q, Ke L, Cai Y (1989) Growth characterization and high titre culture of GCHV. Virol Sin 3:314–339
Fang Q, Attoui H, Cantaloube JF, Biagini P, Zhu Z, de Micco P, de Lamballerie X (2000) Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (genus Aquareovirus, family Reoviridae). Biochem Biophys Res Commun 274:762–766
Fang Q, Seng EK, Ding QQ, Zhang LL (2008) Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch Virol 153:675–682
Fang Q, Sanket S, Liang YY, Zhou ZH (2005) 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China Ser C Life Sci 48:593–600
Kar AK, Bhattacharya B, Roy P (2007) Bluetongue virus RNA binding protein NS2 is a modulator of viral replication and assembly. BMC Mol Biol 8:4
Ke L, Fang Q, Cai Y (1990) Characteristics of a new isolation of hemorrhagic virus of grass carp. Acta Hydrobiol Sin 14:153–159 (in Chinese with English abstract)
Kobayashi T, Ooms LS, Chappell JD, Dermody TS (2009) Identification of functional domains in reovirus replication proteins μNS and μ2. J Virol 83:2892–2906
Mertens PPC, Attoui H, Duncan R, Dermody TS (2005) Reoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier, London, pp 447–454
Miller CL, Arnold MM, Broering TJ, Hastings CE, Nibert ML (2010) Localization of mammalian orthoreovirus proteins to cytoplasmic factory-like structures via nonoverlapping regions of μNS. J Virol 84:867–882
Miller DJ, Schwartz MD, Ahlquist P (2001) Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75:11664–11676
Mohd Jaafar F, Fang Q, Attoui H (2008) Complete characterisation of the American grass carp reovirus genome (genus Aquareovirus: family Reoviridae) reveals an evolutionary link between aquareoviruses and coltiviruses. Virology 373:310–321
Nibert ML, Schiff LA, Fields BN (2001) Reoviruses and their replication. In: Knipe DM, Howley PM (eds) Field virology. Lippincott Williams & Wilkins, Philadelphia, pp 1679–1728
Parker JSL, Broering TJ, Kim J, Higgins DE, Nibert ML (2002) Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol 76:4483–4496
Rangel AAC, Rockemann DD, Hetrick FM, Samal SK (1999) Identification of grass carp hemorrhage virus as a new genogroup of aquareovirus. J Gen Virol 80:2399–2402
Roizman B, Knipe DM (2001) Herpes simplex viruses and their replication. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (eds) Fields virology, 4th edn. Lippincott Williams & Wilkins, Philadelphia, pp 2399–2460
Silvestri LS, Taraporewala ZF, Patton JT (2004) Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. J Virol 78:7763–7774
Touris-Otero F, Cortez-San Martín M, Martínez-Costas J, Benavente J (2004) Avian reovirus morphogenesis occurs within viral factories and begins with the selective recruitment of σNS and λA to μNS inclusions. J Mol Biol 341:361–374
Touris-Otero F, Martinez-Costas J, Vakharia VN, Benavente J (2004) Avian reovirus nonstructural protein μNS forms viroplasm-like inclusions and recruits protein σNS to these structures. Virology 319:94–106
Wei T, Shimizu T, Hagiwara K, Kikuchi A, Moriyasu Y, Suzuki N, Chen H, Omura T (2006) Pns12 protein of rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J Gen Virol 87:429–438
Zhang LL, Lei CF, Fan C, Fang (2009) Expression of outer capsid protein VP5 of grass carp reovirus in E. coli and analysis of its immunogenicity. Virol Sin 24:545–551
Zhang Y, Guo D, Geng H, Liu M, Hu Q, Wang J, Tong G, Kong X, Liu N, Liu C (2007) Characterization of M-class genome segments of muscovy duck reovirus S14. Virus Res 125:42–53
Acknowledgments
We thank Prof. Simon Rayner for critical reading of the manuscript, and Ms. Xiaoyun Sun for assistance in experiments. This work was partly supported by grants from the National Basic Research Program of China (973 Program, 2009CB118701) and the National Natural Scientific Foundation of China (30871940).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fan, C., Shao, L. & Fang, Q. Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol 155, 1755–1763 (2010). https://doi.org/10.1007/s00705-010-0753-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0753-6