Abstract
Aquareoviruses are nonenveloped multiple-shelled particles composed of a genome of 11 double-stranded RNA segments, which encode at least 12 proteins. Among the seven approved Aquareovirus species (Aquareovirus A-G), the striped bass reovirus in the Aquareovirus A (AqRV-A) group and grass carp reovirus (GCRV) in the Aquareovirus-C (AqRV-C) group have been well studied. Intact virions are composed of five inner shell proteins (ISPs; VP1–VP4 and VP6) and two outer capsid proteins (OCPs; VP5 and VP7). The inner core proteins contain all endogenous enzymes responsible for viral transcription and replication. During cellular entry, intermediate sub-viral particles or core particles can be generated by the uncoating of the OCPs VP5 and VP7. In addition to the seven structural proteins of the viral particle, the remaining five proteins have been approved as nonstructural (NS) proteins (NS80/NS1, NS38/NS2, NS31/NS3, NS26/NS4, and NS16/NS5), except the newly identified NS12. Three genotypes of GCRVs (GCRV-I, -II, and -III) have been classified from infected grass carp in China. Studies indicate that NS80 and NS38 of GCRV-I are related to viral inclusion body formation during replication and morphogenesis. Furthermore, NS16 and NS12, encoded by the S7 segment, have been found to play critical roles as fusion-associated small transmembrane proteins to induce syncytium formation via cell–cell fusion. However, no such protein, instead of a fiber protein on the particle surface, which is a counterpart of the orthoreovirus σ1-cell attachment protein, has been found in GCRV-II. Moreover, other NS proteins may be involved in viral replication and pathogenesis via interaction with host cells. This chapter outlines the molecular biology of aquareoviruses.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
2.1 Introduction
Genus Aquareovirus is one of the approved members of the family Reoviridae and grouped in the Spinareovirinae or “turreted” sub-family [30]. The viruses have been isolated from freshwater and seawater aquatic organisms in a wide range of global water areas. Generally, aquareoviruses (AqRVs) are of low pathogenicity in breeding aquatic animals and often detected by routine examination of seemingly healthy fish and shellfish [32]. However, some members of this genus are important fish pathogens that are capable of causing severe hemorrhagic disease, hepatitis, and pancreatitis, leading to a high mortality rate in some cases [12, 32, 33, 37, 52, 54, 60]. Therefore, these viruses pose a significant threat to global aquaculture.
Similar to other members of the Reoviridae family, AqRVs contain 11 double-stranded RNA (dsRNA) genome segments that are packaged into two well-organized concentric protein shells, called the inner or core shell and outer capsid. The viral particles are nonenveloped icosahedral particles with a diameter of approximately 75 nm, which has been determined by negative staining in transmission electron microscopy (TEM) analysis [27, 32, 46, 67]. AqRVs, belonging to the Spinareovirinae group, have relatively large spikes or turret structures situated at the 12 icosahedral vertices of either the intact virion or the core particle [2, 17]. Morphologically, these viruses resemble mammalian orthoreoviruses (MRVs), while the 11 genomic segments are similar in composition to those in the members of the “non-turreted” genus Rotavirus [30]. Based on the morphological and genomic properties of AqRVs isolated from aquatic animals, these were initially referred to as reovirus-like or rotavirus-like agents [36, 46, 54]. However, no cross-reactions are detected between turreted MRVs and non-turreted rotaviruses according to the antigenicity and reciprocal RNA–RNA dot-blot hybridization analyses in earlier studies [9, 32, 38, 52]. Recent nucleotide sequence analysis and comparisons between Aquareovirus and other genera in the family Reoviridae have suggested that there is a close evolutionary relationship between members of the genera Aquareovirus and Orthoreovirus [1, 2, 13], which has been further supported by three-dimensional structural reconstruction studies (in Chap. 3). Seven Aquareovirus species (Aquareovirus A-G) have been approved by the International Committee on Taxonomy of Viruses [30, 39].
Since the first AqRV, golden shiner reovirus (GSRV), reported in the 1970s [46], a large number of AqRVs have been discovered [11, 27, 32, 36, 39, 45, 54, 60, 64, 71], and some pathogenic isolates have complete genome sequences characterized. However, their pathogenic roles in aquaculture remain to be fully elucidated. This chapter summarizes the molecular characteristics of several well-characterized AqRVs, especially grass carp reovirus (GCRV) and striped bass reovirus (SBRV), thus providing a general understanding of the natural biology and molecular properties of AqRVs. For GCRV, the strain GCRV-873 (GCRV-I), originally named as grass carp hemorrhage virus (GCHV), has been suggested as the prototype strain of GCRV species, and hence, it will be referred to as GCRV throughout this chapter.
2.2 Particle-Based Properties
2.2.1 Virus Proliferation and Sample Preparation
Unlike non-fusogenic orthoreoviruses, most AqRV isolates, such as GSRV, American oyster reovirus (strain 13p2), chum salmon reovirus (CSRV), channel catfish reovirus (CCRV), SBRV, GCRV, threadfin reovirus (TFV), and Atlantic halibut reovirus (AHRV), can efficiently replicate in permissive cell lines and induce syncytia as a typical cytopathic effect (CPE). The mature virions can be efficiently released from infected cells by cell lysis; hence, they are fusogenic. Cell lines such as chinook salmon embryo (CHSE), fathead minnow (FHM), bluegill fry (BF-2), Ctenopharyngodon idella kidney (CIK), and others are the most commonly used cell lines for plaque assays and proliferation of AqRVs for viral purification [11, 27, 32, 36, 39, 45, 54, 60, 64, 67, 71]. In addition, SBRV has been reported to replicate well in permissive mammalian cell lines (such as CV-1, HeLa, and Vero) and form typical syncytia [53]. In general, AqRV-infected cell cultures are harvested 3–6 days post-infection (p.i.) or when the CPE is extensive enough for virus purification and identification.
There are many different methods used for AqRV particle isolation and purification from infected cells or diseased fish tissues [16, 52, 67]. Despite their respective advantages in virion isolation, the traditional centrifugation method is commonly used. For general purification, some chemical and physical methods have been used on infected cells in the early stages. Sonication has been used to break virus-infected cells; thus, nascent virus particles can be efficiently released from infected cells. Next, using deoxycholate and Freon extraction, the virus particles are disassociated from the infected cell culture mixtures [52, 67]. In addition, owing to its fusogenic nature, AqRV can be disassociated from cell lysates using physical methods with several freeze-thaw cycles [14,15,16, 54]. After obtaining the virus-cell suspension, centrifugation at several different speeds generally needs to be performed to remove cell debris extensively. The first low-speed centrifugation is used to remove cell debris from the infected culture supernatant, followed by ultracentrifugation to precipitate viral particles. The virus pellet is suspended in 1× SSC (0.15 M NaCl, 0.015 M sodium citrate, pH 7.4) or 1× phosphate-buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.5). To obtain viral components for viral protein analyses or reconstruct three-dimensional images of viral particles, a CsCl equilibrium density or sucrose gradient purification is required for isolating various virus particle components from pelleted virus-cell preparations [15, 16, 67]. After obtaining a uniform viral layer in the centrifugation tube, harvested virions are either extensively dialyzed against TM buffer (50 mM Tris-HCl, pH 7.6, 10 mM MgCl2) or further centrifuged to remove sucrose or salts. The concentrations of the purified viral particles can be determined from optical density measurements at 260 nm, as previously described [15, 16]. For freshly purified virions, short-term storage at 4 °C is the best for further TEM analysis. For long-term storage or maintenance of the structural integrity of purified AqRV virions, storage temperatures of −30 °C or − 80 °C are recommended.
2.2.2 Different Particle Components Extracted from Purified Virus Preparations
Intact virions are the predominant form of mature particles released from infected cell cultures or extracted from aquatic animal tissue homogenates. Intact virions of AqRV can readily undergo particle uncoating to yield intermediate or infectious sub-viral particles (SVPs) and core particles. This phenomenon is consistent with the orthoreoviruses or MRVs, wherein the SVP or the core also appears naturally in the course of infection and plays specific roles in the early and late replication stages [16, 43]. In addition, SVPs or cores can be generated in vitro by protease treatment of purified virions. Chymotrypsin or trypsin is commonly used to convert reovirus virions into infectious SVPs or cores in vitro [16, 35, 40].
MRV virions are relatively stable in natural environments and maintain infectivity for years when stored below 4 °C [43]. Similarly, AqRVs retain relatively high infectivity when stored at or below 4 °C [16, 51]. However, unlike the relatively stable MRV particles, the AqRV SVPs can easily be generated after few days of storage at 22–26 °C or 4 °C in laboratory conditions without undergoing any external treatment with proteases [16, 40]. In most cases, various viral components (including empty and core particles) can be observed from purified sample preparations by TEM.
2.2.2.1 Intact Virions and Empty Particles
During AqRV purification, two viral component layers with a clear opalescent appearance are often obtained after CsCl density gradient centrifugation, which correspond to approximately 1.31 and 1.37 g/cm3 in buoyant density from the top of the tube to the bottom. The upper layer (top component) largely comprises double-shelled empty particles, while the lower layer contains intact virions or native virions (Fig. 2.1, upper panel). Electron micrograph of negatively stained purified virus preparations shows that an AqRV appears as a nonenveloped icosahedral particle with a double-layered capsid and is approximately 75 nm in diameter with 5:3:2 symmetry. The particles viewed along the fivefold axis of symmetry exhibit 20 capsomeres on the periphery of the outer capsid shell. Consistent with MRVs, purification of AqRV particles from infected cells often yields a substantial proportion of particles that lack genomic dsRNA [16, 67]. These empty particles have a lower buoyant density in the CsCl gradient and their respective layer is closer to the top of the gradient in comparison with intact particles (containing dsRNA genome), and hence, the empty particle is termed as the “top component.” Both intact and incomplete (empty) particles are observed in most purified AqRV preparations [11, 14,15,16, 45, 54, 71]. Moreover, the morphology of AqRV particles is strikingly the same as that of MRVs [15, 67]. The top component particles appear similar to intact virions in their morphology, but the central region of these particles can be penetrated by the stain, reflecting the absence of genomic dsRNA in the interior region of the top component particles [16].
Electron micrographs of negatively stained GCRV particles showing various particle components. Intact (left in upper panel) and empty particles (right in upper panel) are isolated from CsCl gradient centrifugation tube (medium in upper panel). Intermediate SVP, intact and empty cores are shown in lower panel. The SVP, intact and empty cores are generated by treatment with 100 mg/ml trypsin at 28 °C for 30 and 120 min, respectively. Double arrowhead in SVP image (left in lower panel) indicates some outer viral capsomeres removed from GCRV particle. Arrowheads indicate pentonal turrets situated at fivefold vertices in intact and empty core images (lower panel). The scale bars represent 100 nm
2.2.2.2 SVP and Virus Core
For a detailed understanding of the particle components of AqRV, different forms of GCRV particles have been isolated from mock-treated and protease-treated virion preparations using an established CsCl gradient centrifugation procedure and then examined under an electron microscope. A visual inspection of the gradient centrifugation showed that the different particles form homogeneous layers with an opalescent appearance [16]. The four distinct layers, including two top component layers, one medium, and one bottom layer, correspond to approximately 1.30, 1.31, 1.38, and 1.44 g/cm3, respectively, in buoyant density from the top of the tube to the bottom. TEM observations revealed that after CsCl gradient centrifugation, the viral top components, including the empty double-layered capsids and empty single core shells, are located in the two top layers, and the intact virions and intact cores stand in the medium and bottom layers, respectively. Here, it should be noted that the intact virion with 1.38 g/cm3 buoyant density might contain a few intermediate SVPs [16], which is very similar to naturally degraded particles of SBRV [40]. This result suggests that parts or some residues of the outermost protein VP7 may fall off from native particles in the virus preparations.
Treatment of AqRVs with proteases (chymotrypsin and trypsin) can generate intermediate SVPs and cores, similar to those reported in other AqRV isolates, such as SBRV [35, 40]. It has been reported that the digestion of the outermost capsid protein (OCP) VP7 of SBRV is associated with increased infectivity [35]. Examination of purified viruses treated with chymotrypsin showed that intact core particles are about 50–60 nm in diameter, which appeared to lack outer capsid capsomeres/proteins in comparison with intact virions of approximately 75 nm. Similarly, infectious SVPs of GCRV can be obtained by treating intact particles with protease (chymotrypsin or trypsin) for a short time. Long protease treatment removes all of the OCPs and exposes prominent turrets, while the core particle structure remains intact (Fig. 2.1, lower panel). Negatively stained TEM images reveal that the core particles exhibit projections at each fivefold vertex and some indistinct projections can be visualized on the empty core shell.
Of note, a few empty single outer capsid shells, empty double outer capsid shells, or single core particles are often found in preparations of natural viral particles without protease treatment, during the purification process or during long-term storage of viral stock at 4 °C. This indicates that the AqRV (or GCRV-873) particle structures are not stable during purification or long-term storage at 4 °C. The related structural architecture of GCRV-873 has been elucidated by three-dimensional image reconstruction [7, 15, 66], and it is described in Chap. 3.
In fact, similar to MRVs, different components of AqRVs (such as SVPs, empty particles, and cores) are also commonly observed in infected cells or tissue sample preparations, except for purified virus preparations that have been treated with chymotrypsin or trypsin under controlled conditions in vitro or virion disassembly during infection of cells [11, 16, 45, 50, 67]. The characteristics and properties of the various AqRV particle components are shown in Table 2.1.
2.2.3 The Virus-Antibody Immunocompound of GCRV
The immunocompound particle of GCRV, which includes its antibody, has been obtained by directly inoculating virions in infected cell culture supernatants with prepared anti-GCRV polyclonal antibodies [14]. In particular, an excellent aggregation of GCRV–antibody immune complexes has been visualized clearly by direct immune electron microscopy after inoculating virus-infected cell supernatants with either anti-VP5 and/or VP7 polyclonal antibody (VP5Ab and VP7Ab) or intact GCRV antibody without performing purification (Fig. 2.2). These results clearly indicate that individually prepared antibodies of outer shell proteins (either VP5Ab or VP7Ab) effectively bind with GCRV particles in cultured cell suspensions, and the binding effect is mostly similar to that of the antibody generated using intact GCRV particles as an antigen. In the case of the control, only a few dispersed particles can be found in GCRV-infected cell supernatants. In fact, the GCRV immunocompound derived from anti-VP7Ab is observed more easily by TEM than that derived from anti-VP5Ab (statistical data for each field not shown here). Furthermore, this is consistent with a previous study showing that enhanced GCRV-neutralizing capacity can be obtained by incubation of GCRV-infected CIK cell culture supernatants with the combination of anti-OCPs (VP5andVP7) Abs [20, 57]. The excellent neutralizing ability of anti-VP5Ab and VP7Ab may be related to the structure and confirmation of the AqRV VP5-VP7 heterodimer complex [7, 15]. It has been reported that the σ1 cell attachment protein of MRV is a major antigen that is able to neutralize reovirus [43]. Despite the lack of σ1 cell attachment protein in GCRV, GCRV VP5 and VP7 proteins represent excellent antigenic epitopes, suggesting that the AqRV VP5 and VP7 proteins have the potential to be used as antigens for vaccines.
2.2.4 Recombinant GCRV Particles
Reoviruses, such as MRV, rotavirus, and bluetongue virus, can automatically assemble into virions by expression of major structural proteins in vitro. As it is known that the AqRV particle is composed of the inner core capsid and the outer shell, an in vitro viral assembly model could be established for AqRVs. Using the baculovirus-based Bac-to-Bac expression system for dual expression of the OCPs VP5 and VP7 or mutant VP5N42A and VP7, which are then combined with purified cores, an in vitro assembly of the recovered native and mutant GCRV intact particles (R-GCRV, R-VP5N42A) has been obtained [70]. Recoated GCRV (R-GCRV, VP5N42A/VP7 R-GCRV) particles closely resemble native GCRV (N-GCRV) in terms of particle morphology and protein composition.
Furthermore, infectivity assays indicate that the recoated particles display infectious properties similar to those of native virions; R-GCRVs with a VP5N42A mutation have been shown to be defective in infectivity and progeny protein expression in infected cells, indicating that a VP5N42A substitution almost completely blocks the autocleavage of the VP5 N-terminus. This observation is consistent with a previous study on MRVs, wherein cores recoated with σ3 and μ1 bearing the N42A mutation have been shown as defective in infectivity [4, 42, 44]. These data indicate that the autocleavage of GCRV VP5 is required for efficient AqRV infection [70].
2.2.5 Surface Labeling of AqRV Particles
Methods of fluorescent labeling of virus particles and cellular structures have been developed to monitor virus trafficking in live infected cells. For enveloped viruses, fluorescent labeling has been successfully applied to trace virus entry and interactions with host cell factors. For nonenveloped viruses, surface coat proteins modified with fluorescent materials can also be realized. Studies have shown that some reoviruses, such as fluorescent-labeled MRV and tetracysteine-tagged bluetongue virus, can retain their biological properties after modification in vitro [76]. To elucidate the AqRV entry into live cells and its interactions with host cell organelles, GCRV virions have been modified in vitro via modification of the surface proteins with biotinylation, followed by conjugation with streptavidin-quantum dots. The GCRV particles modified by quantum dots retain their native biological functions and infectivity. In addition, the quantum dot-labeled GCRV particles show intact, and hence, quantum dots are suitable for use as fluorescent markers to study the molecular mechanisms of viral entry. Moreover, using quantum dot-labeled GCRV, the caveolae/raft-mediated endocytosis viral entry pathway has been identified for efficient AqRV infection [76].
2.2.6 Infectivity Assays of Purified or Modified Virions and SVPs
AqRV infectivity is less affected by particle purification and modification. Studies indicate that the intermediate SVPs of SBRV and GCRV are more infectious than the intact virion and core particle [16, 35]. Several experiments indicated that treatment with 200μg/mL of trypsin or chymotrypsin for approximately 5, 15, or 30 min at 37 °C increases infectivity by more than 2–3 logs in viral titer compared with that of the mock-treated viral particles. However, longer treatments of AqRVs with proteases consistently decrease viral titers because the AqRV core is less infectious, similar to that of the MRV or rotavirus [35], suggesting that AqRVs share common infectivity characteristics with other reoviruses. Plaque assays determining the infectivity of purified and quantum dot-labeled AqRVs have shown that these retain excellent infectivity similar to that of native GCRV [76].
2.3 AqRV Structural Protein Profile
2.3.1 Identification of AqRV Particle Proteins
Following established purification methods, highly purified virions of AqRV have been obtained. The structural protein profiles of AqRVs purified through sucrose gradient show protein components and gel-mobility types similar to those obtained by separating proteins from the purified virion preparation via CsCl gradient centrifugation and analyzed using vertical slab polyacrylamide gels (SDS-PAGE) of 10–15% concentration [15, 16, 52, 54, 67].
CSRV structural proteins were first characterized in 1983 [38]. Five major structural proteins with molecular weights of 137, 126, 72, 44, and 34 kDa have been identified by SDS-PAGE analysis. The two proteins with molecular weights of approximately 72 and 34 kDa have been shown to be present at higher concentrations compared with the other identified proteins. Moreover, SDS-PAGE analysis of purified CSRV has shown five major (132, 130, 68, 43, and 32 kDa) and two minor (110 and 56 kDa) structural proteins [32]. In addition, few minor polypeptides with molecular weights of 110, 94, 80, 62, and 31 kDa have also been detected [32, 67]. In 1984, similar findings were reported for CCRV by Hedrick et al. [21].
In 1987, the first comparative analysis of structural proteins between four AqRV isolates (GSRV, 13p2, CCRV, and CSRV) was performed [67]. Five major structural proteins were compared using SDS-PAGE. Two large polypeptides (approximately 135 and 125 kDa), one medium-sized polypeptide of approximately 70 kDa, and two small polypeptides of 45 and 34 kDa were separated. Of the major structural proteins identified, two proteins with molecular weights of approximately 70 and 34 kDa were consistently present in the highest amount among the four isolates [67]. Minor virion proteins were detected but not characterized. Indeed, due to the limited copy numbers of minor virion proteins in particles, weak bands can be observed sometimes on the stained gel with the naked eye, but are hardly imaged at most times. In addition, SDS-PAGE analysis of isolated landlocked salmon aquareovirus polypeptides revealed the presence of five structural polypeptides with molecular weights ranging from 139 to 32 kDa. The polypeptide separation pattern is similar, but unique when compared with that of GSRV, CSRV, CCRV, and 13p2 [32].
In 1990, five polypeptides with molecular weights ranging from 130 to 35 kDa were identified from purified SBRV [52]. Further, the viral proteins of SBRV have been compared with those of the Atlantic salmon reovirus HBR, Smelt reovirus, Atlantic salmon reovirus (ASV), and turbot reovirus (TRV) by SDS-PAGE [32]. Similar to the identified structural proteins in AqRVs, all five viruses have been shown to contain five major structural proteins (two large, one medium, and two small), with molecular weights ranging from 137 to 34 kDa. The SBRV protein separation pattern is very different from that of SA11 and reovirus type 1 (T1L) [52].
In 1992, the polypeptides of GCRV with molecular weights ranging from 120 to 27 kDa were reported, which were analyzed by SDS-PAGE of purified particles, with separation of 11 polypeptide segments on the gel [28]. In 2002, a comparative analysis of structural proteins between TFV and GCRV revealed that structural proteins with similar molecular weights were present in both the AqRV species [14, 54]. SDS-PAGE of the virion proteins revealed five major structural proteins, approximately 136, 132, 70, 41, and 33 kDa in size. Similarities in the 136-, 132-, and 41-kDa protein bands are observed. The only major difference in the separation pattern between the two species is that with respect to the approximately 70-kDa band in TFV and the approximately 65-kDa band present in GCRV (Fig. 2.3). However, the smallest major protein band was not observed in GCRV in this study [54], which might be because of protein degradation due to the purified GCRV specimen being shipped to Singapore.
In addition, patterns of seven structural proteins have been determined for GCRV-991 with five major and two minor structural proteins [14]. The seven identified structural protein components (VP1–VP7) are approximately 138, 137, 136, 79, 67, 43, and 34 kDa in size, respectively. Three of the seven proteins, VP1, VP2, and VP3, have similar large molecular masses. VP6 and VP7 have relatively small molecular masses. Furthermore, VP5 is the most abundant protein, while VP4 is the least abundant. Similar to the structural protein profiles of GSRV, 13p2, CCRV, and CSRV, minor virion proteins can often be detected but hardly imaged based on the purified virion concentration. The GCRV protein components identified by SDS-PAGE analysis match perfectly with the GCRV structural proteins and their localization in the particle as resolved by three-dimensional image reconstruction of a single particle [7, 15, 16]. Moreover, these results are also consistent with the protein properties of AqRVs and MRVs [34, 67] and predictions deduced from amino acid sequences of GCRV [1, 13].
2.3.2 Various Particle Forms and Related Protein Profiles
It is known as difficult to obtain highly pure virus particles and exactly distinguish the viral particle components from infected cell lysate components upon careful virus purification, unless the virus is labeled with radioactive material during proliferation in infected cells or using western blot analysis with antibodies specific to each viral protein. In addition, avoiding degradation of intact virions during the whole purification process is also a preexisting condition for obtaining highly purified virus particles and their complete particle components. Indeed, the viral particles need to be purified very cautiously because the outermost protein VP7 of the AqRV is readily uncoated in native surroundings or purification processes. Although each step in virion purification is performed carefully at 4 °C, it is very common to observe various particle forms by TEM. Intact AqRV protein components and other viral-type particle components (including the top component, SVP, and inner core) have been identified in GCRV, SBRV, or other AqRV species [16, 32, 35, 40, 67]. For the protein components of GCRV cores (both intact and empty), two major bands are visualized by SDS-PAGE [16]. One large band represents VP1–VP3, while the other represents the VP6 protein. Furthermore, the VP6 band appears very weak in the top empty core sample, suggesting that some of the VP6 molecules in the empty core shell may have been lost during the uncoating of VP5 and VP7 by protease treatment or automatically detached during virus purification. For intact virions and double-shell empty particles, the separated one or two large, one or two medium, and two small protein bands with molecular sizes of 139–132, 70, 43–34, respectively, can be visualized in gel by staining with Coomassie brilliant blue. Of note, in most cases, two bands of the VP5 protein (68 kDa, full-length VP5) and its cleavage segment VP5C (64 kDa) can be detected by SDS-PAGE, which is consistent with the observations for μ1 and cleaved μ1C of MRV [42, 44]. The other cleavage segment VP5N is hardly detectable because it weighs 4 kDa. Consistently, μ1 protein is also mostly found in virions as fragments, μ1N/VP5N (4 kDa) and μ1C/VP5C (72 kDa), which are generated by autocleavage at the site N42-P43, as confirmed by biochemical analysis and three-dimensional imaging at atomic resolution [44, 66]. This suggests that the VP5 protein in the GCRV particle can exist either in two conformations or mainly in the VP5C conformation, as observed in most cases [16, 70]. In addition, since the molecular weights of the identified structural proteins of GCRV are between 34 kDa (VP7) and 136–138 kDa (VP1–VP3), it is not possible to resolve the higher-molecular-weight proteins VP1–VP3 and lower-molecular-weight protein VP7 using the same electrophoretic conditions. Because of the instability of AqRV particles and particle related nature of autocleavage or protease induced cleavage as well as the remaining minor protein components or purification issues with experimental condition limitations, it is not surprising that more than seven structural proteins have been identified in earlier studies [28, 65].
2.4 In Vitro Endogenous Transcription Assays of GCRV Particles
Similar to other reoviruses, the dsRNA genome segments of AqRVs are enclosed by the inner capsid shell. Enzymes that catalyze RNA transcription, capping, and replication are contained within the inner capsid, which serves as the site for the self-RNA synthetic activities. The RNA-dependent RNA polymerase (RdRP)-related endogenous transcription activities of GCRV have been confirmed in earlier studies.
2.4.1 GCRV Nucleic Acid Release
To directly observe the genome of GCRV, the viral RNA component is released using the nucleic acid shadowing method with viral cores being prepared by digesting purified virus with a-chymotrypsin as described previously [28]. The released RNA strand of GCRV is attached to the core turrets (Fig. 2.4a), indicating that the hollow-like spike at the fivefold axes is the channel for RNA release from the inner core into the cytoplasm, which is the same as that in MRVs [3].
Electron micrographs of stained and shadowed preparations of GCRV core and reaction cores. (a, b) GCRV nucleic acid release image of non-transcribing (a) and transcribing GCRV reaction core (b). Few GCRV genome RNA strands released and attached to cores from non-transcribing GCRV core in A2 as arrow indicated, and the non-transcribing core looks loosen after nucleic acid released. Many nascent nucleic acid released from transcribing cores in B. Arrowheads in B2 indicate where strands appear to emerge from spikes, and the transcribing core looks solid after some nascent nucleic acid released. The GCRV nucleic acid release from non-transcribing and transcribing GCRV reaction core preparations is followed as references [22, 28]
2.4.2 GCRV Reaction Core
RNA transcript products in GCRV reaction cores synthesized in vitro have been observed using a combined staining and shadowing method [22]. Up to 12 strands of nascent mRNA have been exhibited in reaction cores by TEM. The reaction cores that are intact and contain attached single-stranded RNA strands have a buoyant density of 1.48 g/cm3 and an A260 to A280 ratio of 1.52. The transcribed genome products of the GCRV reaction cores with [α-32P]-ATP labeling have been detected by analyzing in SDS-PAGE, which are consistent with the full-length copies of their dsRNA templates. An obvious feature of the reaction cores is that the sites of extrusion are probably the 12 hollow projections located on the surface of the core, and many strands are extruded in the form of loops (Fig. 2.4b). This evidence clearly indicates the endogenous transcription activity of GCRV [22].
2.4.3 In Vitro Transcription Assays
The endogenous RdRP activity of various GCRV particles (intact virion, top component, and core) has been assayed by in vitro transcription using [α-32P]-ATP as a marker in earlier studies [23]. Transcriptase activation can be affected by different salt concentrations, temperatures, and reaction times. It has been shown that the cores can be activated for transcription, but the intact GCRV virions are incompetent for producing mRNA. Moreover, the intact GCRV virion can only possess transcriptase activity when the virus sample is resolved in a low-salt buffer, thereby showing effective transcriptase activity. Transcriptase activity assays performed for intermediate SVPs showed SVPs with activity higher than that of the purified viral core particles, suggesting that transcriptase can be activated by uncoating intact GCRV outer capsid shell because the outer capsid shell of intact GCRV can be loosened in a low-salt reaction buffer. Indeed, purified cores possess RNA transcriptase activity, but under the presence of the protective outermost shell, the enzyme activity can be affected. In addition, RdRP is more effective at 28 °C than at a temperature lower or higher than 28 °C, indicating that the optimum temperature for AqRV transcription is 28 °C, as determined by in vitro transcription assays. This is consistent with the optimal replication temperature for GCRV, as demonstrated by in vivo infection and in vitro expression studies of RNA polymerase (VP2) activities [12, 68]. The GCRV RdRP-related transcriptase complex has been verified by recent three-dimensional structural imaging analyses [66].
2.5 The AqRV Genome
The AqRV genome, which is encased by a double-layered protein capsid shell (designated as the inner core and outer capsid), has been shown to be a dsRNA genome due to its resistance to treatment with RNase I and green orthochromatic appearance with acridine orange staining [27, 32, 54, 67]. The genome of AqRVs is composed of 11 dsRNA segments that are packaged in equimolar ratios within a concentrated core. Seven distinct species (Aquareovirus A to Aquareovirus G) have been proposed by the International Committee on Taxonomy of Viruses based on reciprocal RNA–RNA hybridization and nucleotide sequence analyses.
2.5.1 Nomenclature and Electrophoresis Profile of the AqRV dsRNA Genome
The electrophoretic patterns of almost all isolated AqRVs are very similar to each other with three large (segments 1–3), three medium (segments 4–6), and five small segments (segments 7–11). Therefore, the dsRNA genome of AqRVs is generally grouped into three size classes, commonly referred to as the large (S1–S3, about 3.9–3.8 kb), medium (S4–S6, approximately 2.3–2.0 kb), and small (S7–S11, about 1.6–0.9 or 1.6–0.7 kb) classes.
Viral dsRNA of AqRVs has been analyzed by SDS-PAGE using vertical slab gels (10% polyacrylamide gel) immersed in Laemmli’s buffer or 1% agarose gel electrophoresis [2]. Almost all AqRV species show unique electropherotypes in SDS-PAGE. Despite the differences in gel mobility, some similar electrophoretic patterns have been observed among AqRV isolates, e.g., dsRNA genome electrophoretic patterns of GCRV and TFV (Fig. 2.5). However, the RNA profiles of some AqRVs (e.g., SBRV, Atlantic salmon reovirus HBR, Smelt reovirus, and Atlantic salmon reovirus (ASV)) in agarose gels have been observed to be very similar [32]. The differences in the mobility of dsRNA segments in SDS-PAGE or agarose gel electrophoresis depend on the gel properties. In addition, the mobility of dsRNA segments in SDS-PAGE gels depends on both their size and secondary structure, while in agarose gels it depends mostly on their size. In fact, the dsRNA genome of AqRVs is more expediently detected on agarose gels than on SDS-PAGE gels when adequate amounts of dsRNA from virus samples are extracted. In contrast, SDS-PAGE is recommended in cases of lower amounts of AqRV dsRNA genomic material, by staining with silver. Of note, despite the fact that most reoviruses isolated from aquatic animals have 11 dsRNA genomic segments, the genomes of some AqRVs contain 10 and/or 12 segments, as noted for piscine orthoreovirus (PRV) and reoviruses having crab hosts [29, 30], respectively, suggesting a complex and diverse molecular evolutionary course of the AqRV genome.
The segmented genome profiles of GCRVs and TFV. (a) The genomes of GCRV-873 and GCRV-991 were resolved by 7% polyacrylamide gel; M: λ-DNA/EcoRI+HindIII. (b) The genomes of GCRV-873 and TFV were resolved by 1% agarose gel electrophoresis; M: 1 kb DNA ladder. Viral dsRNA genome segments in a and b were visualized following ethidium bromide staining
2.5.2 Genome Sequence-Based Features of AqRVs
The complete genomic sequences of the prototype AqRV strains GSRV and GCRV-873 classified in the species group Aquareovirus-C (AqRV-C) and other AqRV species isolates, such as CSRV and AHRV, have been characterized using the single-primer amplification method-cloned cDNA libraries or an optimized strategy for full-length amplification of cDNA [1, 11, 39, 45, 64, 71]. According to the GenBank data, at least 17 full-length AqRV genome sequences and many partial genome sequences have been deposited thus far. The total length of the AqRV genome sequence is in the range of approximately 23,500–24,500 bp. The lengths of individual gene segments vary from 728 bp (the shortest; S11 gene segment) to 3949 bp (the longest; S1 gene segment), according to the National Center for Biotechnology Information (NCBI). Generally, the GC content of the AqRV genome ranges from 52% to 60%; however, the recently reported AHRV genome consists of 11 segments (with a total size of 24,171 bp) and has a relatively low GC content (ranging from 47.8% to 52.7%) [60]. Most homologous gene segments of different isolates display little or no variation in segment length, such as those of GSRV or GCRV, CSRV, Turbot Scophthalmus maximus reovirus(SMReV), Micropterus salmoides reovirus (MSReV), and SBRV. However, gene segments in the small class vary greatly in length and are more divergent at nucleic acid and protein levels, compared with the large and medium class segments. Generally, each segment encodes one protein, but the S7 segment in the different species groups appears to be bicistronic or tricistronic [2, 30, 73]. Notably, AqRVs are recognized to be fusogenic with syncytia formation as a typical CPE in their permissive cell cultures, which is consistent with the predicted protein functional features of the S7 segment homologous to that of fusogenic avian reovirus (ARV) [8, 24]. However, few AqRV species, such as GCRV-ZH08, GCRV-GD108, and GCRV-104/109, and PRV, show non-fusogenic characteristics [11, 29, 45, 64, 71], which is consistent with that observed for MRVs [43]. The fusogenic and non-fusogenic properties of AqRVs are related to the nature of the S7 segment-encoded proteins.
2.5.3 Terminal Non-coding Regions
The partial and complete genomic sequences of a number of AqRV strains have been determined [1, 2, 11, 39, 45, 47, 48, 55, 56, 64, 71]. In AqRVs, genomic dsRNA segments contain six and five conserved nucleotides at the 5′ terminus and 3′ terminal basic group, respectively (Table 2.2). Particularly, the conserved nucleotide terminal region, with five nucleotides “-UCAUC,” at the 3′ end in AqRVs is same as that in MRV [43], and the extreme 5′ end with six-nucleotides is conserved within a particular virus species. For example, the conserved 5′-and 3′-terminal sequences of isolates in AqRV-C are 5′-GUUAUU……UCAUC-3′, compared to 5′-GUUUUA……UCAUC -3′ in AqRV-A,-B, and –G, and 5′-GUAAUU……UCAUC -3′ or 5′-GUAACU……UCAUC -3′ in unclassified GCRV-ZG08/108 and GCRV-109 species [1, 25, 45, 55, 56, 64, 71]. Similar to other reoviruses, each AqRV gene segment also contains terminal non-coding regions (NCRs) of a particular length. The lengths of the NCRs of genome segments vary across different AqRV species. Generally, the NCRs at the 5′ ends are shorter than that at the 3′ ends in AqRVs. The length ranges from 12 to 35 bp at the 5′ ends, while the 3′ ends contain NCRs of length ranging from 20 to more than 50 or 100 bp depending on the isolated virus strains [25, 45]. Moreover, the first and last nucleotides in the NCRs of all segments are complementary (G-C) and are known to be highly conserved within all AqRV species. Similar to other reoviruses, the NCRs of AqRVs are likely to include sequences important for RNA packaging, recognition by their RdRP for initiating positive- and negative-sense RNA strand synthesis, and translational efficiency [43].
2.5.4 Protein-Coding Assignments and Nomenclature
The coding assignments and properties of the AqRV proteins encoded by each of the 11 genomic segments are now fairly well characterized. The final assignments were determined by in vitro translation using mRNA or genome sequencing using denatured dsRNA combined with three-dimensional structural reconstruction-based methods. It has been confirmed that the 11 genomic segments of AqRVs encode seven structural and five nonstructural (NS) proteins. According to the nomenclature of the viral structural proteins (that are contained in the virion), structural proteins are designated as VP followed by a number, with VP1 being the largest and VP7 being the smallest structural protein. Moreover, the genome segments encode proteins that are not contained in the viral particle, termed as NS proteins. For the NS proteins, two sets of nomenclature are currently used based on either the molecular weight or a number, for example, GSRV NS80 or NS1. The initial assignment of individual proteins to specific gene segments of AqRVs has been performed by in vitro translation of viral mRNAs in early studies on SBRV and GCRV [28, 52, 62, 65]. GCRV polypeptides encoded by each of the gene segments are designated by the molecular weights corresponding to the respective genome segments S1–S11 (Fig. 2.6).
Gene-protein-coding assignment of GCRV. A SDS-PAGE gel shows the 11 segments of dsRNA that comprise GCRV genome and the proteins encoded by each of these genes (a). The genome segments encode nonstructural (NS) and structural (virion) proteins marked at left and right, respectively. The virion proteins are analyzed on a 12% SDS-PAGE gel (b). The viral genome and structural proteins of GCRV are obtained from purified particle
2.5.4.1 SBRV Gene-Protein-Coding Assignment
The gene-protein-coding assignments of SBRV have been well characterized by in vitro translation of individual genome segments [52, 62]. [35S]methionine-labeled SBRV-infected cell lysates were used to analyze the polypeptides and gene-protein-coding assignments of SBRV. Twelve proteins with apparent molecular weights of 130, 127, 126, 97, 73, 71, 46, 39, 35, 29, 28, and 15 kDa were detected in the infected cell lysate, but not in the uninfected cells [32, 52], suggesting that these proteins are encoded by the SBRV genome. Analyzing radiolabeled cell lysates and radiolabeled purified virus proteins using SDS-PAGE and autoradiography for self-exposed X-ray film, seven structural (VP1–VP7) and five NS proteins (NS97, NS39, NS29, NS28, and NS15) have been further classified. There are two major structural proteins, whereas the other five structural proteins are present in smaller amounts. In addition, segments 1–10 of the SBRV genome encode one protein each, while segment 11 encodes two proteins.
To confirm the above data, further translational analysis of the individual genome segments using a nuclease-treated rabbit reticulocyte lysate in vitro was conducted by Subramanian et al. in 1994 [62]. The synthesis of viral proteins by in vitro translation was quantitatively and qualitatively very similar to that observed in SBRV-infected CHSE-214 cells. Of the 12 proteins, the genome segment 11 codes for two proteins, NS29 and NS15. According to the understanding of the proteins encoded by the 11 genome segments of SBRV and their three-dimensional image reconstruction, the proteins VP5 (S5; 71 kDa) and VP7 (S10; 34 kDa) comprise the outer shell [40]. VP1–VP3 and VP6 are the core proteins. Additionally, VP1 is the most likely candidate protein responsible for the turret structure, while the role of VP4 is unknown [40].
2.5.4.2 GCRV Gene-Protein-Coding Assignment
The gene-protein-coding assignment of GCRV was reported by Ke et al. in 1992 [28]. The 11 polypeptides isolated from purified virus particles with molecular weights ranging from 130 kDa to 27 kDa were observed by SDS-PAGE analysis. Furthermore, the denatured viral genome segments were translated using a cell-free rabbit reticulocyte translation system labeled with L-[35S]methionine, which showed 11 protein bands in the exposed X-ray film with autoradiography. Further gene-protein-coding assignment of GCHV was conducted with the wheat germ translation system labeled with L-[35S]methionine using each separated GCRV genome segment [65]. The translation products displayed on the X-ray film showed 12 protein bands with probable molecular weights in the range of 130–19.5 kDa. However, the study failed to distinguish the structural and NS proteins of GCRV. This might be due to poor experimental conditions and some technology-related problems in the 1990s. Indeed, the gene-protein-coding assignments of GCRV were obscure until 2002 [1]. Complete sequence determination and identification of GCRV structural proteins from purified natural virus particles indicated that the GCRV 11 dsRNA genome segments encode 12 proteins. Segments 1, 2, 3, 4, 6, 8, and 10 encode seven structural proteins that form the virus particle [1, 7, 15, 76], and the remaining segments encode five NS proteins that are involved in viral replication and transcription [18, 19, 77, 78].
Further full-length sequence analysis indicated that each dsRNA genome segment of GCRV generally contains a single gene encoding a single protein, except the S7 segment. Twelve proteins have been translated from the 11 gene segments of GCRV, such that 10 of these gene segments encode a single protein, and only the S7 gene encodes two proteins. Genome sequence-based homologous protein analysis indicated that 7 of the 12 proteins are structural components of the virion (VP1–VP7) and five are NS (NS80, NS38, NS31, NS26, and NS16) proteins involved in viral replication [1]. The GCRV particle structural protein compositions were further confirmed by SDS-PAGE, cryo-electron microscopy, and three-dimensional image reconstruction analyses [7, 15, 66]. The 11 segments of dsRNA that comprise GCRV genome and the proteins encoded by each of these genes are shown in Fig. 2.6. Recently, a putative NS12 protein has been identified to be encoded by the S7 genomic segment [73].
2.5.5 In Vitro and In Vivo Protein Expression Analyses
Based on the obtained genome sequence of AqRVs, almost all proteins (including 7 structural and 5 NS proteins) encoded by GCRV genome have been expressed in vitro using different expression vectors (including prokaryotic and yeast expression plasmids, recombinant baculovirus Bac-to-Bac insect expression system, and mammalian expression plasmids) [63, 69, 72]. All the expressed proteins in vitro have been shown to correspond to the predicted protein products from the corresponding GCRV genome, which validates the deduced protein sequences from the corresponding dsRNA genome segment open reading frame (ORF). In addition, the structural and NS protein expressions in virus-infected cells were detected and confirmed with prepared multi-clonal antibodies generated from individual genome segments [69, 76,77,78]. It is important to note that some additional or truncated isoforms of the GCRV proteins, such as VP5/VP5C, NS80/NS80C, which are homologous to MRV protein μ1 and μNSC, respectively, have been detected in both transfected and infected cells [5, 26, 69, 70]. Moreover, NS80 and NS38 expressions are detected prior to that of the structural proteins, indicating that the NS proteins are important for viral replication [69, 78]. As a fusogenic reovirus, the fusion-associated small transmembrane (FAST) protein NS16/NS12, which is similar in function to the ARV p10, p14, and p15 [49], has also been detected in infected and transfected cells. These findings indicate that the AqRV genome may perform translation in infected cells beyond the ATG in-frame rules [25, 49]. The structural proteins and their functions are described in Chap. 3, while the AqRV replication-related NS proteins are described further in this chapter.
2.6 AqRV Morphogenesis
The most characteristic feature of reovirus replication in infected cells is the formation of cytoplasmic factories, named viral inclusion bodies (VIBs), viral factories, or viroplasms, which consist of progeny viruses, usually appearing as paracrystalline arrays of full and empty virions. VIBs are specific intracellular compartments for reovirus replication and assembly [43]. AqRV morphogenesis in host cells has been studied extensively using the ultrastructure and molecular biology methods, which are related to evaluating the mechanism of VIB formation in infected cells [5, 45, 58, 69, 77, 78]. It has been found that the NS proteins NS80 and NS38 of the AqRV as well as the viral core structural proteins play critical roles in AqRV morphogenesis.
2.6.1 TEM Analysis of VIB Formation in Infected Cells
The thin-section ultrastructural TEM images revealed that the AqRV replication and assembly occur within VIBs in infected fish or cultivated cells [32, 51]. Paracrystalline arrays have been observed in cell lines infected by various AqRVs, such as GSRV, 13p2, TRV, GCRV, TFV, TSRV, and AHRV [10, 32, 54, 74]. It has been found that the AqRV particles bind to the plasma membrane of permissive cells at 0 or 10 min post-infection (p.i.) and are subsequently internalized. The SVPs are found in the cytoplasm at 40 and 60 min p.i [51, 76]. It has been shown that the core particles of AqRV are observed within the dense viroplasms in the early stages of infection, and the size and number of viroplasmic inclusions increase as the infection progresses; mature virions are formed with an overall size of about 70–80 nm in the late stage of replication [10, 51]. In addition, a number of viroplasms can be detected at one time in the cytoplasm of a cell, suggesting that there is no limitation on the number of virions penetrating and replicating in a single cell [10, 51].
Once an AqRV species enters the cell, electron-dense particles condense in the periphery of the viroplasms, and the cellular ultrastructural changes can be observed in ultra-thin section images during early infection [10, 32]. The AqRV infection forms neoorganelles in infected cells. The dense globular inclusion bodies/viroplasms appear to be the site for initiating assembly of the viral progeny in the AqRV replication cycle (Fig. 2.7a). The presence of viral polypeptides inside the viroplasms at the medium and late replication stages has been observed by immune electron microscopy and immunofluorescence assays [51, 69, 77, 78]. During replication and assembly of AqRVs, these VIB-like structures have a peculiarly dense consistency that distinguishes them from the adjacent cytoplasm and causes them to appear highly refractile when viewed by phase-contrast microscopy, and they contain fully or partially assembled viral particles, viral proteins, dsRNA, and microtubules [58, 77]. VIBs are formed throughout the cytoplasm of infected cells as the infection progresses. Studies clearly indicate that the AqRV replication is typical to the cytoplasm of infected cells, which is consistent with the reovirus replication features in general.
NS80 and NS38 are involved in viral inclusion formation. (a, right) VIBs in cytoplasm of infected cells (left is the zoomed-in view of the VIB shown in right). (b) Schematic representation of aquareovirus NS80 C-terminal regions with four potential inclusion formation related domains. (c) NS80 induced VIBs were detected in infected and transfected cells. (d) The subcellular localizations of NS80 and NS38 were detected in GCRV-infected CIK cells by IF. The subcellular localizations of NS80 and NS38 immunostained with rabbit anti-NS80 or/and mouse anti-NS38 polyclonal antibodies followed by FITC-conjugated goat anti-rabbit IgG (green) and Texas Red-conjugated goat anti-mouse IgG (red), respectively. Nuclei were counterstained with DAPI (blue). Images in c and d are modified from reference [58]
2.6.2 NS80: A Scaffold of VIBs
The AqRV NS protein NS80, also termed NS1, has been identified as the major constituent for forming globular VIBs [58]. GCRV NS80 consists of 742 amino acids and is encoded by a gene on S4 (2320 nt), with a molecular weight of approximately 80 kDa. Phylogenetic analyses indicated that the NS protein NS80 of GCRV or the homologous protein in other AqRV isolates is related to the formation of VIBs. BLAST analysis comparing the GCRV NS80 analogue with that of CSRV in the genus Aquareovirus and MRV and ARV in the genus Orthoreovirus showed identities mostly within the C-terminal region of the protein. In particular, the NS80 homologous proteins across different AqRV species share two typical coiled-coil regions (513–548 and 615–700 amino acids) in their carboxyl-proximal region (Fig. 2.7b), which have been shown to be important in VIB formation in MRVs [10, 26, 58]. Immunofluorescence assay showed that NS80 can form VIBs when expressed alone in transfected cells or during viral infection (Fig. 2.7c). It has been also identified that the C-terminus of NS80 is responsible for VIB formation, and the N-terminal NS80 interacts with viral proteins [58, 77]. In addition, a shorter specific fragment of NS80, an approximately 58-kDa product of NS80, has been detected in infected and transfected cells [5]. As NS80 is a multi-functional protein, the different isoforms of NS80 proteins present in infected cells may be functional at various replication stages, suggesting that the isoforms may play different functions during viral replication.
2.6.3 Interaction of NS80 with Viral Proteins
It has been observed that NS80 retains five ISPs (VP1–VP4 and VP6) and NS38 within VIBs in co-transfected or infected cells (Fig. 2.7d). NS80 interacts with each core protein and newly synthesized viral RNAs colocalized with VIBs [26, 69, 77]. Furthermore, time-course analysis of the viral structural protein expression showed that the expression of NS80 is first detected, followed by that of the ISP VP3 and other ISPs, suggesting that VIBs are essential for the formation of progeny virions. Further experiments indicated that knockdown of NS80 by shRNA not only inhibits the expression of the AqRV structural proteins, but also reduces viral infection. These results indicate that NS80-based VIBs are formed at an earlier stage of viral infection, and the protein NS80 is able to coordinate the expression of viral structural proteins and viral replication [69, 77].
2.6.4 Role of NS38 in GCRV Morphogenesis
AqRV NS38, encoded by GCRV S9 (1130 nt), contains 352 amino acids with a molecular mass of approximately 38 kDa. BLAST analyses suggested that there is approximately 23% similarity between the AqRV NS38 and MRV σNS at the protein level. It has been found that protein σNS of MRVs, together with another NS protein μNS, and protein σ3 can associate with mRNA molecules to form single-stranded RNA-containing complexes [43, 78]. Some studies have implicated σNS as a minimally essential viral component for forming VIBs together with μNS, which then recruits other reovirus proteins and RNA to initiate viral genome replication. Studies have indicated that the AqRV NS38, a virus genome-encoded putative single-stranded RNA-binding protein, interacts with not only NS80 in VIBs, but also ISPs (VP1–VP4 and VP6). Interactions between NS38 and NS80-RNA complexes in both transfected and infected cells have also been detected. Knockdown of NS38 by siRNAs-115/219 clearly reduces viral infection, with decreased mRNA and protein yields. Moreover, NS38 can interact with the host cellular eukaryotic translation initiation factor 3 subunit A (eIF3A) in transfected cells. Furthermore, it has been identified that NS38 directly interacts with outer shell protein VP5 and VP7 (unpublished data). These findings indicate that NS38 may function as a mediator by interacting with the viral and host cellular components in VIBs during viral replication and particle assembly [78].
2.7 GCRV S7 Genome Segment-Encoded FAST Protein and Associated Syncytium Formation
Similar to fusogenic orthoreoviruses that can promote the formation of multinucleated syncytia in infected cells [8, 49], AqRVs are able to induce typical CPEs and finally lead to multinucleated syncytium formation in permissive cell lines during cultivation. AqRVs and fusogenic orthoreoviruses are the only known examples of nonenveloped viruses that lead to cell–cell fusion and syncytium formation in virus-infected cells [24, 49]. Studies have indicated that the FAST protein encoded by the AqRV S7 genome segment is responsible for cell–cell fusion during the viral replication cycle.
2.7.1 Detection of FAST Protein
Early investigations have revealed that ARVs can induce multinucleated syncytia formation in infected cell cultures, which distinguishes them from non-fusogenic MRVs [49]. It has also been found that the fusogenic nature of ARVs is such that the formation of cellular syncytia is not related to viral cellular entry or exit, as is the case for enveloped viruses, such as the human immunodeficiency virus (HIV) gp41 (HIV-1), which mediates fusion between target cells through a complex interaction of viral glycoproteins with cell receptors [24]. FAST proteins are small (95–198 amino acids) and expressed as NS proteins during the viral replication cycle [49]. Studies on the reovirus fusion mechanism suggest that the reovirus FAST proteins may function as virus genome-encoded fusogens and utilize many accessory host factors to drive cell–cell fusion, and therefore, may benefit the rapid spread of infection after being expressed in infected cells [8, 49]. A recent study on ARVs demonstrated that cell–cell fusion induced by the reovirus FAST proteins enhances replication and pathogenicity of nonenveloped dsRNA viruses [24].
2.7.2 Sequence-Based Features of FAST Protein
Fusogenic reovirus-induced cell–cell fusion and syncytium formation are dependent on a distinct family of FAST proteins encoded by polycistronic genomic segments. Some FAST proteins of the ARV, such as p10, p14, and p15, have been well characterized in the genus Orthoreovirus [8, 24]. A few FAST proteins, such as the p22 protein from AtSRV, SMReV, and MSReV (AqRV-A) and NS16 from GCRV and GSRV (AqRV-C), have been predicted in the genus Aquareovirus based on bioinformatics analyses [18, 25, 49]. Subsequently, the proteins NS16, NS22, or p22 encoded by the polycistronic genome segment S7 of AtSRV, MSReV, SMReV, and GCRV have been identified as FAST proteins in the genus Aquareovirus [6, 18, 25]. Generally, the basic structure of the FAST protein (Fig. 2.8a), including the transmembrane (TM) domain, polybasic (PB) region, hydrophobic region, and polyproline and proline-rich motif, has been determined in the AtSRV p22, GCRV NS16, and MSReV/SMReV NS22 [6, 18, 25, 49]. In particular, sequence analysis has also suggested an additional dileucine motif in the cytoplasmic region at amino acid positions 113–114, which may regulate protein sorting and other cellular processes.
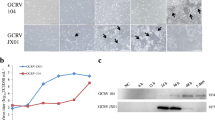
Identification of GCRV genome S7 encoded FAST protein. (a) Diagram of AqRVs genome segment S7 encoded proteins NS16/NS22, NS12, and NS31/NS32. Virus strains and accession numbers for segment 7 of the genomes are as follows: AqRV-A: MsReV (KJ740731.1), SMReV (HM989936.1); AqRV-C: GCRV-873 (AF403393.1), GSRV (AF403404.1). (b) Expression of GCRV S7 encoded NS16 and NS31 protein. NS16 induced cell–cell fusion and multinucleated syncytium formation in transfected CIK cells (left), no syncytium formation is observed with NS31 expression (right). Cells were fixed at 48 hpt and stained with HE or Hoechst. (c) Cytopathic effects are observed in GCRV-infected CIK cells at 12 hpi. Mock-infected cells served as a negative control. (d) Syncytia formation in transfected CIK cells with recombinant plasmids (pCI-Neo) containing different truncation regions of GCRV S7 sequence. (e) Immunofluorescence assays of truncated S7 fragment expressions in transfected cells. The expressed proteins were detected by IF assay at 24 hpt with an anti-FLAG MAb followed by Alexa 488 (green) or 568 (red) labeled secondary antibody, respectively. Arrows indicate the syncytia and nucleus aggregation
Bioinformatics analysis has shown that NS16 and NS22 of the AqRV share basic structural motifs with the ARV FAST proteins p10, p14, and p15. NS16 and NS22 in AqRV-C and AqRV-A species, respectively, have been predicted to be single-pass membrane proteins comprising 146 and 198 amino acids, respectively, which display essential features similar to those of the identified FAST protein motifs and are assumed to possess an N-terminus-outside/C-terminus-inside asymmetric topology on the plasma membrane. In the N-terminal region, there is a strong TM domain (covering 37–60 amino acids for NS16 and 35–57 amino acids for NS22). Following the TM domain, there is a PB region that contains a stretch of basic residues (covering 63–78 amino acids for NS16 and amino acid positions 61–68 and 82–95 for NS22). The PB region is thought to support the translocation of the N-terminal domain (34 amino acids for NS22 and 36 amino acids for NS16) into the extracellular environment [18], and it has been implicated as essential for FAST protein activity. In addition to the TM domain, a hydrophobic region called the hydrophobic patch has been predicted to exist in the C-terminal fragment of both proteins (amino acid positions 113–121 for NS16 and 140–150 for NS22). Moreover, two regions rich in arginine, proline, and histidine have been found in NS22 [25]. In contrast, there are three regions rich in arginine, proline, and histidine in the S7 encoded FAST protein of the AtSRV [49], despite its high sequence similarity with the SMReV NS22. Furthermore, some differences have been found in the motifs in NS22 and NS16. A myristoylation consensus sequence (MGXXXS) has been identified at the N-terminus of NS22 [25], but no such myristoylation site exists in NS16 of GCRV or GSRV. It has been reported that N-terminal myristoylation is necessary for the fusion activity of the reptilian reovirus protein p14 [8, 41]. Notably, the viruses utilize a noncanonical CUG codon for initiating translation to produce the fusion protein p22 responsible for syncytiogenesis [25, 49, 73].
Interestingly, GCRV-104 initially appeared to lack an NS FAST protein. However, further careful analysis indicated that the potential N-terminal region of NS15 (ORF2) in the S11 segment may contain a TM domain with its size consistent with that of the FAST protein ectodomains; additionally, a cluster of basic residues contained at the C-terminus that is enriched in arginine and proline residues, and several cysteine residues that may be palmitoylated [41]. Similarly, a bioinformatics assay predicted that the GCRV-HZ08 NS41 and/or GCRV-GD108 NS11/9 (ORF2) potentially encode membrane-interacting NS proteins having one or more TM domains [41]. These may be additional examples of non-fusogenic integral membrane proteins encoded by AqRVs that are similar to the PRV protein p13 [29]. However, these predicted transmembrane proteins with relative biological functions are not clearly defined in AqRVs and orthoreoviruses.
2.7.3 NS16 and NS22: The AqRV FAST Proteins
Bioinformatics analysis has shown that NS16 and NS22 (p22 for the AtSRV) of the AqRV share basic structural motifs with the reovirus FAST proteins, suggesting that the AqRV NS proteins NS16/NS22 may be a fusion protein responsible for AqRV-C and AqRV-A syncytiogenesis. As expected, the expression of NS16 (ORF14-454 nt) in transfected cells could induce cell–cell fusion and multinucleated syncytium formation (left panel in Fig. 2.8b), consistent with typical CPE formed in CIK cells infected with GCRV (Fig. 2.8c). However, the expression of S7 cDNA with another ORF (GCRV NS31: 520–1344 nt; SMReV NS32: 489–1325 nt) does not cause cell–cell fusion (right panel in Fig. 2.8b). Especially, many large syncytia formed when full-length cDNA of the S7 segment expressed in transfected CIK cells at 24 hpt, hinting that one more membrane fusion relevant ORF contained in the 5′ end sequence of S7 segment. In fact, detailed analyses have shown that cell–cell fusion and nucleus aggregation could be formed when truncated constructions of S7 fragment, such as S7(1-454), S7(1-518), expressed in transfected cells (Fig. 2.8d,e), indicating that the initial 5′ end ORFs of the GCRV and SMReV/MSReV S7 segment encode proteins critical for cell–cell fusion [6, 25]. It may need to note that the NS16 alone is capable of inducing cell–cell fusion and syncytium formation in transfected cells; nevertheless, the fusion activity is less efficient in comparison with the viral infection [18]. Unlike NS16, which has an AUG ORF, NS22 of SMReV has been found to be translated from a non-AUG translation start site. In addition, the NS16 FAST protein has been observed to display an N-terminus-outside/C-terminus-inside orientation, with the N-terminal ectodomain critical for effective fusion [18]. Moreover, immunofluorescence assays of GCRV have revealed that NS16 co-localizes with the NS protein NS26 in co-transfected cells. The enhanced fusion efficiency can be detected when NS16 is co-expressed with NS26, implying that NS26 may participate in cell–cell fusion through cooperation with NS16 in AqRV infection [18, 19].
2.7.4 NS12: A Novel Membrane-Associated Protein
It is known that the GCRV S7 segment encodes two proteins NS16 and NS31. A neglected ORF, tentatively named NS12, residing between NS16 and NS31 in the S7 segment, has been found to have atypical fusogenic activity [73]. With an additional ORF(195-518), the nucleotide sequence of NS12 partially overlaps with the 3′ expressible nucleotide sequence of NS16 (Fig. 2.8a). Furthermore, bioinformatics analysis has indicated that NS12 is a transmembrane protein, which has been confirmed by its exclusive presence in the membrane-associated fraction of the cell lysate. In particular, the NS12 product can be detected in infected cells, indicating that NS12 is expressed in cells with GCRV infection. However, the expression of NS12 alone cannot induce visible syncytium formation in transfected CIK cells (Fig. 2.8e), which distinguishes it from the approved FAST protein NS16. Subsequent bioinformatics analysis showed that NS12-like ORFs (with an AUG or non-AUG initiation codon) are also present in the S7 segment of other AqRVs [73], suggesting that NS12 homologs may be widely distributed in the genus Aquareovirus. Collectively, the novel membrane-associated protein NS12 is functionally distinct from the known AqRV FAST protein NS16. The identification of bicistron/tricistron mRNAs in AqRVs and their genetic variety provides a basis for further understanding the mechanism of molecular evolution of fusogenic AqRVs.
2.7.5 Role of NS31 in Virus Replication
Compared with the aforementioned AqRV NS proteins, very little is known about the role of NS31 or NS32, which is encoded by the GCRV or SMReV/MsReV genomic segment S7 downstream of NS16 or NS22 (Fig. 2.8a). When NS31or NS32 solely expressed in transfected cell, no syncytium formation can be observed [25] (right panel in Fig. 2.8b). Immunofluorescence assays showed that NS31 colocalized with NS16 in transfected cells (unpublished data), hinting that NS31 might associate with NS16 during cell–cell fusion. However, no biological relationship has been determined between the GCRV S7 segment-encoded FAST proteins NS16 and NS31. Bioinformatics analysis has predicted that the NS31 protein contains a helix-turn-helix-like domain and a C-terminal acidic α-helix motif. Using a GAL4-based yeast reporter system and a grass carp cDNA library, it has been found that a fusion protein composed of the Gal4-BD domain and NS31 (BD-NS31) is able to activate the expression of reporter genes (Gal1/MEL1 promoter) without the Gal4-AD domain. In addition, NS31 homologues from other AqRVs have been shown to possess a similar transcriptional activation in yeast, suggesting that the AqRV/GCRV NS31 protein may be a potent transcription regulator. Further analyses are found that GCRV NS31 and other NS31-like proteins in Aquareovirus genus could efficiently induce host heat-shock 70-kda protein (HSP70) expression, and GCRV protein synthesis or progeny virus yields was restrained by an inhibitor of host HSP70 in infected cells. All these results indicated NS31 cognate proteins in the Aquareovirus genus should play a regulating role during aquareovirus replication [72].
2.7.6 Possible Function of GCRV NS26 in Syncytia Formation
GCRV NS26 is encoded by the S11 genomic segment, which shares no sequence homology with the known proteins in MRVs. NS26 has been found to interact with NS80 and co-localize with NS16 in co-transfected cells. Co-expression of NS16 with NS26 has been observed to enhance cell–cell fusion efficiency [18]. These results suggest that NS26 may participate in the fusion process of viral infection. Further functional analysis of NS26 has suggested that the TLPK motif is important for NS26 to enhance the fusogenic activity of NS16, and NS26 may utilize lysosomes to benefit the fusion activity [19].
2.8 MRV σ1-like Cell Attachment Protein of GCRV-II Species
In contrast to the majority of AqRVs, the newly identified GCRV-ZH08, GCRV-GD108, and GCRV-104/-109 have been found to have the MRV σ1-like cell attachment protein (VP55 or VP56) on the particle surface [11, 41, 45, 64, 71]. The recombinant MRV σ1 protein of GCRV-GD108 can bind to grass carp snout fibroblast cells, as observed by cell attachment assays. Neutralization tests have shown that the polyclonal antibody generated from the expressed fiber protein VP55 is able to prevent viral infection in both fish and grass carp snout fibroblast cells. A more recent study indicated that fiber-like protein VP55 can repress interferon production by degrading the phosphorylated cellular transcription factor interferon regulatory factor 7 (IRF7) [75]. As σ1 protein is a major epitope and possesses hemagglutination activity, further studies on the GCRV-ZH08/-GD108/-104/-109 (GCRV-II) S7 segment-encoded cell attachment protein will provide a basis for understanding the mechanism of VP55 in cell entry and pathogenesis.
2.9 Conclusions and Future Considerations
According to classic gene-protein-coding assignment and genome sequence-based protein prediction, the 11 dsRNA genome segments of GCRV (AqRV-C) encode at least 12 proteins. Of the 12 approved proteins, seven structural proteins VP1–VP7 have currently been confirmed by high-resolution single-particle three-dimensional image reconstruction, as well as SDS-PAGE analysis of the structural proteins of the viral particle. The basic biological and molecular characteristics of the remaining proposed NS proteins NS80, NS38, NS31, NS26, and NS16 have also been investigated [5, 10, 18, 25, 26, 58, 69, 72, 78]. In addition, a novel protein NS12, encoded by the S7 genomic segment of GCRV, has recently been identified to play a partial role in syncytium formation [73]. This evidence suggests that another unknown NS protein may be expressed in virus-infected host cells and facilitate AqRV replication. Much progress has been made in the elucidation of the AqRV molecular biology in the past 30 years. However, for more in-depth knowledge, some issues need to be addressed in future studies.
Particle Instability of AqRVs
A number of morphological studies have shown that various particle components (intact virions, SVPs, empty particles, and inner cores) can be observed in highly purified or partially purified AqRV preparations by TEM. Indeed, based on the fusogenic properties of most AqRVs inoculated in permissive host cells, mature virions can be released from infected cells by lysis. For general purification of the GCRVs, the collected virus-cell suspension can be purified directly without undergoing any physical methods, such as sonication, or chemical treatment, such as treatment with deoxycholate and extraction with freon, which are frequently used in the purification of the non-fusogenic Orthoreovirus [16, 43, 61]. A combination of freeze-thaw cycles before conducting differential centrifugation or ultracentrifugation might be a routine preparatory procedure used in the AqRV purification to obtain a high yield of virus from infected cell lysates. Despite the lack of protease treatment in the general purification of GCRV from lysed cellular components, some degraded viral particles can be observed in untreated viral sample preparations, suggesting that the GCRV particle is capable of degrading spontaneously without the action of exogenous proteases. The fact that there are some SVPs present in the purified viral stock leads us to speculate that the AqRV particle capsid structures are not stable or outer shell protection protein VP7 can easily become detached from penetration protein VP5 under natural conditions or during storage. Based on this phenomenon, stringent and careful handling of viral preparations is needed during AqRV purification. The unstable nature of the AqRV particles might be related to the host-dependent conditions beneficial for establishing efficient infection.
Gene-Protein-Coding Assignment and Protein Nomenclature
Generally, each protein of AqRVs encoded by a genome segment is named by its corresponding size. The coding assignments of the SBRV genome segments were the first to be well determined among the identified AqRVs by in vitro translation in 1994 [32, 62]. Comparison of [35S]methionine-radiolabeled proteins from infected cell lysates and purified virions has led to the distinction of the structural and NS proteins [52, 62]. In this way, 12 proteins (with molecular weights of 130, 127, 126, 97, 73, 71, 46, 39, 35, 29, 28, and 15 kDa) that are encoded by the 11 genomic segments have been identified. Each SBRV segment is monocistronic, with the exception of segment 11, which encodes two NS proteins. Furthermore, of the 12 proteins, seven are structural proteins: VP1 (~130 kDa), VP2 (~127 kDa), VP3 (~126 kDa), VP4 (~73 kDa), VP5 (~71 kDa), VP6 (~46 kDa), and VP7 (~35 kDa) [32, 59, 62], and the remaining five are NS proteins. Further studies on SBRV with SVP-enhanced infectivity assays and single-particle three-dimensional images have shown that segments S5 and S6 encode proteins VP4 and/or VP5, respectively, which resemble the protein product μ1 in MRV encoded by the M2 genome segment [40]. VP4 and VP5 may be isoforms of protein VP5, as reported by McPhillips et al. [35]. The VP5 protein appears to be generated from the products of “VP4,” which correspond to the homologous MRV proteins μ1 and μ1C [44]. In fact, comparing the native virion and 5 minut trypsinized [5MT] structures of SBRV shows a noticeable structural change in the trimeric subunits [40]. This observation is highly correlated with the biochemical data of SBRV infectivity assays performed by treatment with protease and visualized by SDS-PAGE [35, 62]. It has been shown that the putative VP5 protein is cleaved after 5 min of trypsin treatment, resulting in a 52-kDa fragment that stays associated with the particle [35]. Further, three-dimensional image reconstruction has been used to determine the VP5 density in an SBRV particle [40]. Therefore, in this regard, the protein VP4 encoded by the S6 segment of the SBRV [35, 62] should be termed as VP5, although no further studies have been conducted on SBRV. Similarly, the copy number of the VP4 protein encoded by the GCRV S5 segment is low, and it is very difficult to detect the protein from purified viral preparations via SDS-PAGE. Early studies have failed to define GCRV gene-protein-coding assignments accurately [28, 64]. Nonetheless, the appropriate gene-protein-coding assignment of GCRV has been obtained and confirmed from purified GCRV proteins by combining full-length genome sequence analysis and three-dimensional image reconstruction [1, 13, 15]. In fact, in another study on MRV [31] and the GCRV S6 sequence [48], the S6 segment-encoded protein has also been defined as VP5. Therefore, based on the current understanding of the molecular virology and high-resolution structural biology of GCRV or AqRVs, it is time to modify the nomenclature of the GCRV-873 (GCRV-I) S6 genome segment-encoded VP4 protein to VP5 to avoid further confusion between VP4 and VP5, which are encoded by genome segments S5 and S6, respectively, in AqRVs.
Proteins Involved in AqRV Morphogenesis
The GCRV NS80, a protein homologous to the μNS protein of MRV, has been well characterized [5, 10, 26, 58, 69, 77, 78]. Studies have shown that the NS protein NS80 of GCRV can form VIBs in singly expressed or infected cells and recruit all the ISPs (VP1–VP4 and VP6) in addition to NS38 within its VIBs [69, 78]. It has been confirmed that the coiled-coil motifs in the C-terminal regions of NS80 are crucial for forming VIBs in infected cells, whereas the N-terminal regions of NS80 play important roles in interacting with viral proteins and supporting viral replication [58, 77]. Similar to σNS in MRV, NS38 is considered to have single-stranded RNA-binding ability and is thought to be involved in viral protein synthesis. In addition to NS80, the GCRV NS38 has also been found to interact with five viral core structural proteins, RNA, and host eIF3A during viral replication for efficient viral protein synthesis. Furthermore, recent evidence showed that NS38 interacts with the OCPs VP5 and VP7 in transfected and infected cells, suggesting that NS38 plays a significant role in progeny particle assembly via interactions with viral proteins and host cell factors. The detailed roles of the viral structural and NS proteins in AqRV morphogenesis remain obscure; therefore, it is further necessary to elucidate the function of each AqRV genome-encoded protein and the interaction among all the proteins during AqRV morphogenesis.
Fusogenic Characteristics of AqRVs
Based on the currently characterized genome sequences and deduced protein functions related to the presence or absence of cell fusion-related NS FAST or surface structural σ1-like cell attachment protein, the genus Aquareovirus should be subdivided into two distinct subgroups relative to the fusogenic and non-fusogenic orthoreoviruses. The evidence for their phylogenetic classification based on this distinct feature is dependent on the diversity of the S7 genomic segment of the AqRVs. In the genus Orthoreovirus, this differentiation is determined by the S class gene, especially the S1 gene in most cases. In fact, most orthoreoviruses, except MRV, are found to encode FAST proteins responsible for inducing cell–cell fusion and syncytium formation [8, 49]. Along with the proteins encoded by fusogenic orthoreoviruses, which are well-characterized examples of transmembrane fusion proteins encoded by nonenveloped viruses, the AqRV FAST proteins are the other known examples. In contrast, the newly identified PRV with 10 genomic segments and the GCRV-II or -III (GCRV-ZH08, GCRV-GD108, and GCRV-104/-109) species have been found to possess a fiber-like cell attachment protein that is similar to σ1, the cell adsorption protein of MRV [41]. Interestingly, it appears that GCRV-104 can induce a typical CPE in infected permissive cells 5 days p.i [11]. This phenomenon may be related to the potential membrane-associated protein NS15 (ORF2), which contains a FAST protein-specific TM domain in the N-terminal region that is found to be encoded by the GCRV-104 genome S11 segment. If this is the case, then GCRV-104 might not only possess σ1-fiber protein in the particle, but also bear fusogenic activity in cell cultures. In addition, a novel membrane-associated protein NS12 has been found to be encoded by the GCRV-I S7 gene segment residing between the gene segments encoding NS16 and NS31. Therefore, this indicates that the fusogenic nature is too complex in AqRVs. To understand the molecular mechanism underlying the pathogenesis of GCRV and other AqRVs, it is important to conduct further extensive and in-depth investigations of the NS FAST-related proteins and their roles in AqRV replication.
To summarize, the molecular characteristics of AqRVs have been extensively studied in recent years. Our current knowledge of the AqRV particle, genome, and encoded protein functions provides a strong basis for further understanding the detailed molecular mechanism of action of AqRV pathogenesis in host cells during infection. Studies on the interactions between NS80 and NS38 and the OCPs VP5 and VP7 as well as the five core proteins during morphogenesis and particle assembly and those on different fusogenic mechanisms used by AqRVs to enhance viral replication efficiency are warranted. In addition, understanding the role of other unknown NS proteins, such as NS26 and NS31, in regulating viral replication in cooperation with host cellular factors is also critical to reveal the detailed events involved in AqRV replication and pathogenesis.
Abbreviations
- AHRV:
-
Atlantic halibut reovirus
- AqRV:
-
Aquareovirus
- ARV:
-
Avian reovirus
- BF-2:
-
Bluegill fry
- CCRV:
-
Channel catfish reovirus
- CHSE:
-
Chinook salmon embryo
- CIK:
-
Ctenopharyngodon idella kidney
- CoSRV:
-
Coho salmon reovirus
- CPE:
-
Cytopathic effect
- CSRV:
-
Chum salmon reovirus
- FAST:
-
Fusion-associated small transmembrane
- FHM:
-
Fathead minnow
- GCHV:
-
Grass carp hemorrhage virus
- GCRV:
-
Grass carp reovirus
- GSRV:
-
Golden shiner reovirus
- ISPs:
-
Inner shell proteins
- MRV:
-
Mammalian orthoreovirus
- MSReV:
-
Micropterus salmoides reovirus
- NCR:
-
Non-coding region
- NS:
-
Nonstructural
- OCPs:
-
Outer capsid proteins
- ORF:
-
Open reading frame
- p.i.:
-
Post-infection
- PB:
-
Polybasic
- PRV:
-
Piscine orthoreovirus
- RdRP:
-
RNA-dependent RNA polymerase
- SBRV:
-
Striped bass reovirus
- SMReV:
-
Turbot Scophthalmus maximus reovirus
- SVP:
-
Sub-viral particle
- TEM:
-
Transmission electron microscopy
- TFV:
-
Threadfin reovirus
- TM:
-
Transmembrane
- TRV:
-
Turbot reovirus
- VIBs:
-
Virus inclusion bodies
References
Attoui H, Fang Q, Jaafar FM, Cantaloube JF, Biagini P, de Micco P, de Lamballerie X (2002) Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, grass carp reovirus, striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae). J Gen Virol 83:1941–1951
Attoui H, Mertens PPC et al (2012) Reoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, pp 541–637
Bartlett NM, Gillies SC, Bullivant S, Bellamy AR (1974) Electron microscopy study of reovirus reaction cores. J Virol 14:315–326
Chandran K, Walker SB, Chen Y, Nibert ML (1999) In vitro recoating of reovirus cores with baculovirus-expressed outercapsid proteins μ1 and σ3. J Virol 73(5):3941–3950
Chen Q, Zhang J, Zhang F, Guo H, Fang Q (2016) Identification and characterization of two cleavage fragments from the Aquareovirus nonstructural protein NS80. Virol Sin 31:314–323
Chen ZY, Gao XC, Zhang QY (2015) Whole-genome analysis of a novel fish reovirus (MsReV) discloses aquareovirus genomic structure relationship with host in saline environments. Viruses 7:4282–4302
Cheng L, Fang Q, Shah S, Atanasov IC, Zhou ZH (2008) Subnanometer resolution structures of the grass carp reovirus core and virion. J Mol Biol 382:213–222
Ciechonska M, Duncan R (2014) Reovirus FAST proteins: virus-encoded cellular fusogens. Trends Microbiol 22:715–724
Dopazo CP, Toranzo AE, Samal SK, Roberson BS, Baya A, Hetrick FM (1992) Antigenic relationships among rotaviruses isolated from fish. J Fish Dis 15:27–36
Fan C, Shao L, Fang Q (2010) Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol 155:1755–1763
Fan Y, Rao S, Zeng L, Ma J, Zhou Y, Xu J, Zhang H (2013) Identification and genomic characterization of a novel fish reovirus, Hubei grass carp disease reovirus, isolated in 2009 in China. J Gen Virol 94:2266–2277
Fang Q, Ke LH,Cai YQ (1989) Growth characteristics and high titer culture of grass carp hemorrhage virus (GCHV)-873 in vitro. Virol Sin (In Chinese with English Abstract)
Fang Q, Attoui H, Biagini JF, Zhu Z, de Micco P, de Lamballerie X (2000) Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (genus aquareovirus, family reoviridae). Biochem Biophys Res Commun 274:762–766
Fang Q, Xiao TY, Ding QQ (2002) Virological properties of GCRV 991 strain. Virol Sinica 17(2):183–187. (In Chinese with English Abstract)
Fang Q, Shah S, Liang Y, Zhou ZH (2005) 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China Ser C 48:593–600
Fang Q, Seng EK, Ding QQ, Zhang LL (2008) Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch Virol 153:675–682
Francki RIB, Fauquet CM, Knudson SL, Brown F (1991) Classification and nomenclature of viruses, 5th report of the international committee on taxonomy of viruses. Arch Virol 2(Suppl):186–1991
Guo H, Sun X, Yan L, Shao L, Fang Q (2013) The NS16 protein of aquareovirus-C is a fusion-associated small transmembrane (FAST) protein, and its activity can be enhanced by the nonstructural protein NS26. Virus Res 171:129–137
Guo H, Chen Q, Yan L, Zhang J, Yan S, Zhang F, Fang Q (2015) Identification of a functional motif in the AqRV NS26 protein required for enhancing the fusogenic activity of FAST protein NS16. J Gen Virol 96:1080–1085
He Y, Xu H, Yang Q, Xu D, Lu L (2011) The use of an in vitro microneutralization assay to evaluate the potential of recombinant VP5 protein as an antigen for vaccinating against grass carp reovirus. Virol J 8:132
Hedrick PR, Rosemark R, Atonstein D, Winton JR (1984) Characteristics of a new reovirus from channel catfish (Ictalurus punctatus ). J Gen Viral 65:1527–1534
Huang J, Ke LH, Cai YQ (1992) Study on the reaction core of grass carp hemorrhage virus. Acta Biochem Biophys Sinica 24(2):132–139
Huang J, Ke LH, Cai YQ (1992) Ribonucleic acid transcriptase activity associated with purified grass carp hemorrhage disease. Chinese J Virol 8(1):50–56
Kanai Y, Kawagishi T, Sakai Y, Nouda R, Kobayashi T (2019) Cell–cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses. PLoS Pathog 15(4):e1007675
Ke F, He LB, Pei C, Zhang QY (2011) Turbot reovirus (SMReV) genome encoding a FAST protein with a non-AUG start site. BMC Genomics 12:323
Ke F, He LB, Zhang QY (2013) Nonstructural protein NS80 is crucial in recruiting viral components to form aquareoviral factories. PLoS One 8:e63737. https://doi.org/10.1371/journal.pone.0063737 PMID: 23671697
Ke L, Fang Q, Cai Y (1990) Characteristics of a new isolation of hemorrhagic virus of grass carp. Acta Hydrobiol Sin 14:153–159. (in Chinese with English Abstract)
Ke LH, Wang W, Fang Q, Cai YQ (1992) Studies on the in vitro translation of grass carp hemorrhage virus and its proteins. Chinese J Virol (8)2:169–193
Key T, Read J, Nibert ML, Duncan R (2013) Piscine reovirus encodes a cytotoxic, non-fusogenic, integral membrane protein and previously unrecognized virion outer-capsid proteins. J Gen Virol 94:1039–1050
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2012) Virus taxonomy, ninth report of the international committee on taxonomy of viruses. Elsevier, San Diego
Liemann SK, Chandran K, Nibert ML (2002) Structure of the reovirus membrane-penetration protein, mu1, in a complex with is protector protein, sigma3. Cell 108(2):283–295
Lupiani B, Subramanian K, Samal SK (1995) Aquareovirus. Annu Rev Fish Dis 5:175–208
Marshall SH, Samal SK, McPhillips TH, Moore AR, Hetrick FM (1990) Isolation of a rotavirus from smelt, Osmerus mordax (Mitchell). J Fish Dis 13:87–91
McCrae MA, Joklik WK (1978) The nature of the polypeptides encoded by each of the 10 double-stranded RNA segments of reovirus type 3. Virology 89:578–593
McPhillips TH, Dinan D, Subramanian K, Samal SK (1998) Enhancement of aquareovirus infectivity by treatment with proteases: mechanism of action. J Virol 72:3387–3389
Meyers TR (1979) A reo-like virus isolated from juvenile American oysters (Crassostrea virginica). J Gen Virol 43:203–212
Meyers TR (1980) Experimental pathogenicity of reovirus 13p2 for juvenile American oysters Crassostrea virginica (Gmelin) and bluegill fingerlings Lepomis macrochirus (Rafinesque). J Fish Dis 3:187–201
Meyers TR (1983) Serological and histopathological responses of rainbow trout, Salmo gairdneri Richardson, to experimental infection with the 13p2 reovirus. J Fish Dis 6:277–292
Mohd-Jaafar F, Goodwin AM, Merry G, Fang Q, Cantaloube J, Biagini P, De-Micco P, Mertens P, Attoui H (2008) Complete characterisation of the American grass carp reovirus genome (genus Aquareovirus: family Reoviridae) reveals an evolutionary link between aquareoviruses and coltiviruses. Virology 373:310
Nason EL, Samal SK, Venkataram Prasad BV (2000) Trypsin induced structural transformation in aquareovirus. J Virol 74:6546–6555
Nibert ML, Duncan R (2013) Bioinformatics of recent aqua- and orthoreovirus isolates from fish: evolutionary gain or loss of FAST and fiber proteins and taxonomic implications. PLoS One 8:e68607
Nibert ML, Fields BN (1992) A carboxy-terminal fragment of protein u1/u1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol 66:6408–6418
Nibert ML, Schiff LA, Fields BN (2001) Reoviruses and their replication. In: Knipe DM, Howley PM (eds) Fields virology. Lippincott Williams and Wilkins, Philadelphia, pp 1679–1728
Odegard AL, Chandran K, Zhang X, Nibert ML (2004) Putative autocleavage of outer capsid protein u1, allowing release of myristoylated peptide u1N during particle uncoating, is critical for cell entry by reovirus. J Virol 78(16):8732–8745
Pei C, Ke F, Chen ZY, Zhang QY (2014) Complete genome sequence and comparative analysis of grass carp reovirus strain 109 (GCReV-109) with other grass carp reovirus strains reveals no significant correlation with regional distribution. Arch Virol 159:2435–2440
Plumb J, Bowser PR, Grizzle JM, Mitchell AJ (1979) Fish viruses: double-stranded RNA icosahedral virus from a North American cyprinid. J Fish Res Board Can 36:1390–1394
Qiu T, Lu RH, Zhang J, Zhu ZY (2001a) Complete nucleotide sequence of the S10 genome segment of grass carp reovirus (GCRV). Dis Aquat Org 44:69–74
Qiu T, Lu RH, Zhang J, Zhu ZY (2001b) Molecular characterization and expression of the M6 gene of grass carp hemorrhage virus (GCHV), an aquareovirus. Arch Virol 146:1391–1397
Racine T, Hurst T, Barry C, Shou J, Kibenge F, Duncan R (2009) Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site. J Virol 83(11):5951–5955
Rangel AAC, Rockemann DD, Hetrick FM, Samal SK (1999) Identification of grass carp haemorrhage virus as a new genogroup of aquareovirus. J Gen Virol 80:2399–2402
Rivas C, Noya M, Cepeda C, Bandin I, Barja JL, Dopazo CP (1998) Replication and morphogenesis of the turbot aquareovirus (TRV) in cell culture. Aquaculture 160:47–62
Samal SK, Dopazo CP, McPhillips TH, Baya A, Mohanty SB, Hetrick FM (1990) Molecular characterization of a rotavirus-like virus isolated from striped bass (Morone saxatilis). J Virol 64:5232–5240
Samal SK, McPhillips T H, Dinan D (1998) Lack of restriction of growth for aquareovirus in mammalian cells[J]. Arch Virol 143(3):571–579
Seng EK, Fang Q, Chang SF, Ngoh GH, Qin QW, Lam TJ, Sin YM (2002) Characterisation of a pathogenic virus isolated from marine threadfin fish (Eleutheronema tetradactylus) during a disease outbreak. Aquaculture 214:1–18
Seng EK, Fang Q, Sin YM, Lam TJ (2005a) Molecular characterization of a major outer capsid protein encoded by the threadfin aquareovirus (TFV) gene segment 10 (S10). Arch Virol 150:2021–2036
Seng EK, Fang Q, Sin YM, Lam TJ (2005b) Molecular cloning, DNA sequence analysis, and expression of cDNA sequence of RNA genomic segment 6 (S6) that encodes a viral outer capsid protein of threadfin aquareovirus (TFV). Virus Genes 30:209–221
Shao L, Sun X, Fang Q (2011) Antibodies against outer-capsid proteins of grass carp reovirus expressed in E. coli are capable of neutralizing viral infectivity. Virol J 8:347
Shao L, Guo H, Yan LM, Liu H, Fang Q (2013) Aquareovirus NS80 recruits viral proteins to its inclusions, and its C-terminal domain is the primary driving force for viral inclusion formation. PLoS One 8:e55334. https://doi.org/10.1371/journal.pone.0055334
Shaw AL, Samal S, Subramanian K, Prasad BVV (1996) The structure of aquareovirus shows how different geometries of the two layers of the capsid are reconciled to provide symmetrical interactions and stabilization. Structure 15:957–968
Skoge RH, Nylund A, Solheim K, Heidrun Plarre H (2019) Complete genome of Atlantic halibut reovirus (AHRV) associated with mortality in production of Atlantic halibut (Hippoglossus hippoglossus) fry. Aquaculture 509:23–31
Smith RE, Zweerink HJ, Joklik WK (1969) Polypeptide components of virions, top component, and cores of reovirus type 3. Virology 39:791–810
Subramanian K, McPhillips TH, Samal SK (1994) Characterization of the polypeptides and determination of genome coding assignments of an aquareovirus. Virology 205(1):75–81
Wang H, Yu F, Li J, Lu L (2016) Laminin receptor is an interacting partner for viral outer capsid protein VP5 in grass carp reovirus infection. Virology 490:59–68
Wang Q, Zeng W, Liu C, Zhang C, Wang Y, Shi C, Wu S (2012) Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J Virol 86:12466
Wang W, Cai YQ, Fang Q, Liu Y, Ke L (1994) Gene-protein-coding assignments of grass carp hemorrhage virus genome RNA species. Virol Sin 9(4):356–361
Wang X, Zhang F, Su R, Li X, Chen W, Chen Q, Yang T, Wang J, Liu H, Fang Q, Cheng L (2018) Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly. Proc Natl Acad Sci 115:7344–7349
Winton JR, Lannan CN, Fryer JL, Hedrick RP, Meyers TR, Plumb JA, Yamamoto T (1987) Morphological and biochemical properties of four members of a novel group of reoviruses isolated from aquatic animals. J Gen Virol 68:353–364
Yan L, Liu H, Li X, Fang Q (2014) The VP2 protein of grass carp reovirus (GCRV) expressed in a baculovirus exhibits RNA polymerase activity. Virol Sin 29:86–93
Yan L, Zhang J, Guo H, Yan S, Chen Q, Zhang F, Fang Q (2015) Aquareovirus NS80 initiates efficient viral replication by retaining core proteins within replication-associated viral inclusion bodies. PLoS One 10:e0126127
Yan S, Zhang J, Guo H, Yan L, Chen Q, Zhang F, Fang Q (2015) VP5 autocleavage is required for efficient infection by in vitro recoated aquareovirus particles. J Gen Virol 96:1795–1800
Ye X, Tian YY, Deng GC, Chi YY, Jiang XY (2012) Complete genomic sequence of a reovirus isolated from grass carp in China. Virus Res 163(1):275–283
Yu F, Wang LL, Li WJ, Wang H , Que SJ, Lu LQ (2019) Aquareovirus NS31 protein serves as a specific inducer for host heat shock 70-kDa protein. J Gen Virol. https://doi.org/10.1099/jgv.0.001363
Yu F, Wang LL, Li WJ, Lu L (2019) Identification of a novel membrane-associated protein from the S7 segment of grass carp reovirus. J Gen Virol. https://doi.org/10.1099/jgv.0.001223
Zainathan SC, Carson J, Nowak BF (2017) Preliminary characterization of Tasmanian aquareovirus (TSRV). Arch Virol 162:625–634
Zhang C, Lu LF, Li ZC, Xiao-Yu Zhou XY, Zhou Y, Chen DD, Li S, Zhang YA (2020) Grass carp reovirus VP56 represses interferon production by degrading phosphorylated IRF7. Fish Shellfish Immunol 99:99–106
Zhang F, Guo H, Zhang J, Chen Q, Fang Q (2018) Identification of the caveolae/raft-mediated endocytosis as the primary entry pathway for aquareovirus. Virology 513:195–207
Zhang J, Guo H, Chen Q, Zhang F, Fang Q (2016) The N-terminal of aquareovirus NS80 is required for interacting with viral proteins and viral replication. PLoS One 11:e0148550
Zhang J, Guo H, Zhang FX, Chen QX, Chang MX, Fang Q (2019) NS38 is required for aquareovirus replication via interaction with viral core proteins and host eIF3A. Virology 2019(529):216–225
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Fang, Q., Zhang, J., Zhang, F. (2021). Molecular Biology of Aquareoviruses. In: Fang, Q. (eds) Aquareovirus. Springer, Singapore. https://doi.org/10.1007/978-981-16-1903-8_2
Download citation
DOI: https://doi.org/10.1007/978-981-16-1903-8_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-1902-1
Online ISBN: 978-981-16-1903-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)