Abstract
A real-time multiplex polymerase chain reaction (rtm-PCR) assay was developed and optimized to simultaneously detect three viral pathogens of shrimp in one reaction. Three sets of specific oligonucleotide primers for white spot syndrome virus (WSSV), infectious hypodermal and haematopoietic necrosis virus (IHHNV) and Taura syndrome virus (TSV), along with three TaqMan probes specific for each virus were used in the assay. The rtm-PCR results were detected and analyzed using the Light Cycler 2.0 system. Forty-five PCR-positive samples and four negative samples were used to confirm the sensitivity and specificity of the rtm-PCR. The rtm-PCR identified and differentiated the three pathogens. With one viral infection of shrimp, a specific amplified standard curve was displayed. When samples from shrimp infected with two or three pathogens were analyzed, two or three specific standard curves were displayed. The sensitivity of the rtm-PCR assay was 2,000, 20, and 2,000 template copies for WSSV, IHHNV and TSV, respectively. No positive results (standard curves) were displayed when nucleic acid from Vibro spp., and Streptococcus spp. DNA were used as PCR templates. The results indicate that real-time multiplex PCR is able to detect the presence of and differentiate each pathogen in infected shrimp. This real-time multiplex PCR assay is a quick, sensitive, and specific test for detection of WSSV, IHHNV and TSV and will be useful for the control of these viruses in shrimp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
White spot syndrome virus (WSSV), infectious hypodermal and haematopoietic necrosis virus (IHHNV) and Taura syndrome virus (TSV) are three major viral pathogens that infect penaeid shrimp [2–4, 6, 8]. Mixed infections with these viruses have been described [12] and can cause high mortality, leading to economic losses that are detrimental to the shrimp farming industry [9, 16, 21].
Multiple diagnostic methods such as histologic examination, electron microscopy and in situ hybridization are required to detect and differentiate these viral pathogens [8, 9, 16, 18]. However, these methods are time consuming and labor intensive. Molecular assays, such as DNA probes [13, 15] and PCR methods, have been used for rapid and sensitive detection of these viruses [7, 10, 14, 19, 20]. Recently we developed multiplex reverse transcription-PCR for the simultaneous differentiation of three viral pathogens of penaeid shrimp [22]. Real-time PCR is preferred over conventional PCR in clinical laboratories because there is no need for post-amplification handling, leading to faster analysis and reduced risk of amplicon contamination [11, 17]. Real-time PCR can also provide an estimate of pathogen titer [1]. Recently, real-time PCR assays have been developed for the separate detection of WSSV, TSV and IHHNV [5, 23]. This study describes a real-time multiplex PCR assay for simultaneous detection of these three viruses.
Materials and methods
Clinical samples and plasmids
WSSV-, IHHNV- and TSV-infected clinical tissue samples and positive controls consisted of recombinant plasmids containing specific and conserved genes of the three viruses (TSV-pMD18-T, WSSV-pMD18-T, IHHNV-pMD18-T). These three plasmids were cloned using the specific primers described previously [22] and are listed in Table 3.
Isolation of nucleic acids from clinical tissues samples
RNA and DNA extractions from WSSV, IHHNV, and TVS isolates were carried out using Trizol according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Total nucleic acid was extracted from clinical tissue samples from shrimp infected with WSSV, IHHNV, and TVS as well as from disease-free white shrimp and Penaeus orientalis Kishinouye according to a method described previously [22]. Extracted DNA of Vibro spp. and Streptococcus spp. were kindly provided by the China Institute of Veterinary Drug Control, Beijing.
Oligonucleotide primers and DNA probes
Three sets of primers and DNA probes, listed in Table 1, were designed to amplify highly conserved gene sequences of WSSV (AF369029), IHHNV (AF218226) and TSV (NC003005) (GenBank sequence data). All three sets of primers and probes were synthesized at TaKaRa, Dalian, China.
Real-time multiplex PCR assay
Amplification reactions were performed in volumes of 20 μl with TaKaRa premix, 40 μM MLV reverse transcriptasease, 16U RNase inhibitor, 0.6 μM primer for WSSV and TSV or 0.4 μM primer for IHHNV, 0.4 μM probe for WSSV and TSV or 0.2 μM probe for IHHNV, and 2 μl of the DNA/RNA sample. The PCR amplification consisted of 2 min at 95°C, followed by 40 cycles of 10 s at 95°C and 30 s at 60°C. Amplification, detection, and data analysis were performed with the Light Cycler 2.0 system (Roche Molecular Biochemicals, Mannheim, Germany).
Sensitivity and specificity of rtm-PCR
The sensitivity of the rtm-PCR was determined using tenfold dilutions of template of each specific plasmid containing specific genes of viruses (WSSV-pMD18-T, IHHNV-pMD18-T and TSV-pMD18-T). The results of the rtm-PCR assay were compared to those of our previously developed PCR assay [22]. To determine the specificity of the rtm-PCR assay, specific DNA fragments from WSSV, IHHNV and TSV were amplified and cloned in pMD18-T cloning vector according to the manufacturer’s protocol (Takara Dalian, China). These three recombinant plasmids were sequenced, and sequence data were analyzed using Dnastar software and compared with the corresponding sequence data in GenBank. For negative controls, DNA from Vibrio and Streptococcus and distilled water were included.
Interference assay and reproducibility
Various concentrations of plasmid containing WSSV, IHHNV and TSV genes (104, 108 and 108 copies of each; or 108, 101 and 108 copies of each; or 108, 108 and 104 copies of each) were mixed together and subjected to rtm-PCR. The copy numbers of the genes were calculated according to the following formula: (copies/μl = (The plasmid’s concentration × 6×1014)/(The plasmid’s size in base pairs × 324.5).
Detection of clinical samples
DNA from 15 clinical samples each from WSSV, IHHNV and TSV that were known to be positive by routine PCR were also subjected to real-time multiplex PCR.
Results
The real-time multiplex PCR assay for detection of three viral pathogens of penaeid shrimp was designed as a multiplex assay for simultaneous detection of WSSV, IHHNV and TSV. Since the presence of other oligonucleotides and fluorescent probe could alter the efficiency of PCR amplification, each set of primers and probe was tested in an individual format as well as in a multiplex format, using different concentrations of the three viruses. The result of these experiments indicates that there was no systematic deviation in the amplification curves when comparing the multiplex assay with the single-target assays. No difference in amplification efficiency was observed between the singleplex and multiplex formats, as measured by the slopes of amplification curves during the exponential phase and the cycle threshold (CT) values obtained with individual samples. Furthermore, the detection limits for the multiplex and individual assay formats were nearly identical, since even the most diluted samples were detected in both types of assay.
Detection limit
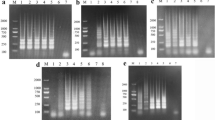
The limit of detection for the real-time multiplex assay was determined with TSV-pMD18-T, WSSV-pMD18-T and IHHNV-pMD18-T that were serially diluted tenfold. The sensitivity of the real-time multiplex PCR assay was 20,000 for WSSV, 20 for IHHNV and 20,000 for TSV template copies, respectively (Fig. 1a–c), and its sensitivity was 10, 1,000 and ten times higher than that of the routine PCR. Standard curves are shown in Fig. 2a–c. Different concentrations of WSSV, IHHNV and TSV, when mixed together, still could be identified by this assay, which implies that the rtm-PCR assay can be used for simultaneous detection of infection with the three viruses.
a Sensitivity of the real-time multiplex PCR for IHHNV. 1 2 × 107 copies/μl; 2 2 × 106 copies/μl; 3 2 × 105 copies/μl; 4 2 × 104 copies/μl; 5 2 × 103 copies/μl; 6 2 × 102 copies/μl; 7 2 × 101 copies/μl; 8 2 × 100 copies/μl; 9 Negative control. b Sensitivity of the real-time multiplex PCR for TSV; 1 2 × 107 copies/μl; 2 2 × 106 copies/μl; 3 2 × 105 copies/μl; 4 2 × 104 copies/μl. c Sensitivity of the real-time multiplex PCR for WSSV. 1 2 × 107 copies/μl; 2 2 × 106 copies/μl; 3 2 × 105 copies/μl; 4 2 × 104 copies/μl
Reproducibility and specificity
The samples were examined repeatedly using the rtm-PCR (Table 2), and the results indicated that the rtm-PCR was reproducible. The rtm-PCR results of different samples showed that one specific amplification curve was displayed when shrimp were infected by only one of these three viral pathogens, whereas two or three specific amplification curves were displayed when shrimp were infected by two or three viral pathogens, and no amplification curves were displayed for samples containing Streptococcus, Vibrio and water (Figs. 2, 3a–c). The results indicate that rtm-PCR was able to detect and differentiate the presence of each pathogen in clinically infected shrimp.
Clinical samples
Forty-five samples that were positive for each single virus by routine PCR were detected by real-time multiplex PCR (Table 3). The results showed that 1.82 × 107–3.34 × 105 copies/μl of WSSV were detected from 15 WSSV samples, and one sample was WSSV and IHHNV positive (7.72 × 106 and 2.10 × 102 copies/μl); 2.13 × 107–8.49 × 103 copies/μl of IHHNV were detected from 15 IHHNV samples, and one sample was both IHHNV and WSSV positive (6.78 × 106 and 1.12 × 104 copies/μl); 1.16 × 107–5.49 × 105 copies/μl of TSV were detected from 15 TSV samples, and none were IHHNV or WSSV positive.
Discussion
The rtm-PCR assay described here uses PCR primers and TaqMan probes targeting conserved regions of WSSV, IHHNV and TSV genes. One main advantage of this assay compared to other available tests is that it is multiplex. By using this approach, it was possible to identify all three pathogens in the same reaction vessel. The simultaneous detection of WSSV, IHHNV and TSV is especially useful, because these viruses commonly cause mixed infection in shrimp [9, 12, 16, 21].
In addition to its use in clinical diagnostics, the multiplex assay may be of value for detection of pathogen-free shrimp in environmental samples. However, the multiplex feature of this assay is optional; if so preferred, the three components can be utilized as single-targeting assays or combined into duplex assays without impacting the quality of the results. This makes this assay adaptable to circumstances that may not require the simultaneous detection of all three for a diagnostic decision.
Another important aspect of this real-time PCR approach is the short turnaround time. The confirmatory result of a suspected WSSV, IHHNV and TSV infection was obtained within 5 h of receiving the sample in the laboratory. This included time for DNA extraction and multiplex real-time PCR assay. The current methods for laboratory diagnosis of these three viruses are labor-intensive and may lack the sensitivity and speed required to reveal the cause of infection before it is too late to take the appropriate measure. The real-time multiplex PCR assay presented here can therefore be extremely useful as a fast and sensitive complement to existing diagnostic methods. In addition, this real-time multiplex PCR does not require the unique expertise involved in morphology-based tests but can be performed in any laboratory with adequate infrastructure for real-time PCR testing.
References
Bell AS, Ranford-Cartwright LC (2002) Real-time quantitative PCR in parasitology. Trends Parasitol 18:337–342
Bonami JR, Hasson K, Mari J, Poulos B, Lightner DV (1997) Taura syndrome of marine penaeid shrimp: characterization of the viral agent. J Gene Virol 78:313–319
Bonami JR, Trumper B, Mari J, Brehelin M, Lightner DV (1990) Purification and characterization of the infectious hypodermal and haematopoietic necrosis virus of penaeid shrimps. J Gen Virol 71:2657–2664
Brock JA (1997) Special topic review: Taura syndrome. A disease important to shrimp farms: in The Americas. World J Microb Biotech 13:415–418
Dhar AK, Roux MM, Klimpel KR (2001) Detection and quantification of Infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using Real-time quantitative PCR and SYBR green chemistry. J Clin Microb 39:2835–2845
Erickson HS, Zarain-Herzberg M, Lightner DV (2002) Detection of Taura syndrome virus (TSV) strain differences using selected diagnostic methods: diagnostic implications in penaeid shrimp. Dis Aquat Organ 52:1–10
Kiatpathomchai W, Boonsaeng V, Tassanakajon A, Wongteerasupaya C, Jitrapakdee S, Panyim S (2001) A non-stop, single-tube, semi-nested PCR technique for grading the severity of white spot syndrome virus infections in Penaeus monodon. Dis Aquat Organ 47:235–239
Lightner DV, Redman RM (1998) Shrimp diseases and current diagnostic methods. Aquaculture 164:201–220
Lightner DV (1996) The penaeid shrimp viruses IHHNV and TSV: epizootiology, production impacts and role of international trade in their distribution in the Americas. Revues Scientifique Et Technique Office International Des Epizooties 15:579–601
Lo CF, Leu JH, Ho CH, Chen CH et al (1996) Detection of baculovirus associated with white spot syndrome (WSBV) in penaeid shrimps using polymerase chain reaction. Dis Aquat Org 25:133–141
Mackay IM (2004) Real-time PCR in the microbiology laboratory. Clin Microbiol Infect 10:190–212
Manivannan S, Otta SK, Karunasagar I, Karunasagar I (2002) Multiple viral infections in Penaeus monodon shrimp postlarvae in an Indian hatchery. Dis Aquat Org 48:233–236
Mari J, Bonami JR, Lightner DV (1998) Taura syndrome of penaeid shrimp:cloning of viral genome fragments and development of specific gene probes. Dis Aquat Org 33:11–17
Nunan LM, Poulos BT, Lighter DV (1998) Reverse transcription polymerase chain reaction (RT-PCR) used for the detection of Taura syndrome virus (TSV) in experimentally infected shrimp. Dis Aquat Org 34:87–91
Nunan LM, Lightner DV (1997) Development of a non-radioactive gene probe by PCR for detection of white spot syndrome virus (WSSV). J Virol Meth 63:193–201
Plumb JA (1997) Trends in freshwater fish disease research, p 35–47. In: Flegel TW, MacRae IH (eds) Diseases in Asian aquaculture III. Fish health section, Asian fish soc, Manila. FAO Aquaculture Newsletter no. 16. FAO, Rome, 28 pp
Silva AJ, Pieniazek NJ (2003) Latest advances and trends in PCR-based diagnostic methods. In: Dionisio D (ed) Textbookatlas of intestinal infections in AIDS. Springer-Verlag Italia, Milan, pp 397–412
Takahashi Y, Itami T, Kondo M, Maeda M, Fujii R, Tomonaga S, Supamattaya K, Boonyaratpalin S (1994) Electron microscopic evidence of bacilliform virus infection in Kuruma shrimp (Penaeus japonicus). Fish Pathol 29:121–125
Tang KFJ, Wang J, Lightner DV (2004) Quantitation of Taura syndrome virus by real time RT-PCR with TaqMan assay. J Virol Meth 115:109–114
Wang SY, Hong C, Lotz JM (1996) Development of a PCR procedure for the detection of Baculovirus penaei in shrimp. Dis Aquat Org 25:123–131
Wang YC, Lo CF, Chang PS, Kou GH (1998) White spot syndrome associated virus (WSSV) infection in cultured and wild decapods in Taiwan. Aquaculture 164:221–231
Xie Z, Pang Y, Deng X, Tang X, Liu J, Lu Z, Khan MI (2007) A multiplex RT-PCR for simultaneous differentiation of three viral pathogens of penaeid shrimp. Dis Aquat Organ 76:77–80
Yue ZQ, Liu H, Wang W, Lei ZW, Liang CZ, Jiang YL (2006) Development of real-time polymerase chain reaction assay with TaqMan probe for the quantitative detection of infectious hypodermal and hematopoietic necrosis virus from shrimp. J AOAC Int 89:240–244
Acknowledgments
This work was supported by the Guangxi Science and Technology and Guangxi Aquaculture and Animal Husbandry Bureau. The authors thank Dr. Herbert Van Kruiningen for his critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xie, Z., Xie, L., Pang, Y. et al. Development of a real-time multiplex PCR assay for detection of viral pathogens of penaeid shrimp. Arch Virol 153, 2245–2251 (2008). https://doi.org/10.1007/s00705-008-0253-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-008-0253-0