Abstract
PERVs are integrated in the genome of all pigs. Some of them infect human cells and represent therefore a potential risk for xenotransplantation using pig cells or organs. Three replication-competent subtypes have been described, PERV-A, PERV-B and PERV-C. Whereas PERV-A and PERV-B are polytropic viruses and infect, among others, human cells, PERV-C is an ecotropic virus, infecting only pig cells. Recombinant PERV-A/C are able to infect human cells, they are characterised by high-titre replication and their proviruses have been found de novo integrated in the genome of somatic pig cells, but not in the germ line. This review compares recombinant PERVs with other recombinant retroviruses in order to evaluate their potential pathogenicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xenotransplantation using porcine cells or organs has been proposed to alleviate the shortage of human donor organs for allotransplantation [1], but it may be associated with the risk of transmission of zoonotic microorganisms. Whereas transmission of most porcine microorganisms may be prevented by designated-pathogen-free (DPF) breeding of the animals, porcine endogenous retroviruses (PERVs) cannot be eliminated in this way [2]. PERVs are integrated into the genome of all pigs, and while they behave like normal cellular genes in that they are inherited by the offspring, they may also be transcribed and translated, resulting in the expression of viral proteins and the release of viral particles and therefore pose a potentially high risk for xenotransplantation [3]. PERVs belong to the genus Gammaretrovirus, and they are closely related to feline leukaemia virus (FeLV), murine leukaemia virus (MuLV), gibbon ape leukaemia virus (GaLV) and koala retrovirus (KoRV), all of which induce leukaemia and immunodeficiency in the infected host [4] (Fig. 1). More than one hundred proviral copies of PERV are integrated in the pig genome, depending on the pig breed, among them three different replication-competent subtypes, PERV-A, PERV-B and PERV-C [5–8]. Most proviruses are defective and unable to produce replication-competent viruses. Whereas PERV-A and PERV-B are present in the genome of all pigs, PERV-C is not ubiquitous. PERVs have been found to be released from normal pig cells [9–14] as well as from pig tumour cell lines [15–18]. In contrast to PERV-A and -B, which infect human cells (human-tropic viruses), PERV-C infects only pig cells (ecotropic virus) [10, 19–23]. PERV-A and PERV-B are polytropic viruses infecting cells from humans, cats, minks and non-human primates, whereas cells from rats, mice, rabbits and cotton rats could not be infected [10, 22–26]. When the receptors for PERV-A were identified [27], it was soon shown that, in mice, a single amino acid mutation is responsible for resistance to PERV infection [28]. In contrast, the receptor was functional in rats; however, its expression appeared to be under a threshold level for supporting PERV-A infection. Overexpression of the receptor finally resulted in infection of rat cells. The tropism of PERVs is determined by two sequences in the receptor-binding domain (RBD) in the surface envelope protein, variable region (VR) A and VRB (Fig. 2). These sequences are different in all three viruses. A third region that shows some variability, the proline-rich region (PRR) is also required for binding to cells, in addition to VRA and VRB. The sequences of all other viral genes, with the exception of the long terminal repeat (LTR), are closely related in PERV-A, PERV-B and PERV-C.
a Schematic presentation of the envelope protein of PERVs, SU surface envelope protein, TM transmembrane envelope protein, VRA virus receptor domain A, VRB virus receptor domain B, PRR proline-rich region. b PERV-A, c PERV-C, d PERV-A/C recombinant found in animals 13910 and 15149, e in animals 13653 and 15150 [55] and f PERV-A/C 50 described in refs. [9, 10, 49]
Recombinations of other retroviruses including HIV-1
Recombination plays an important role in the evolution of retroviruses. Recombination between highly related viruses reassorts sequences, thereby increasing the diversity of the population and allowing the emergence of variants that are fit for the particular selection pressures at a given time. Recombination can also occur between genetically similar viruses that are further apart in sequence homology than those in a viral population, for example, recombination between different subtypes of human immunodeficiency virus type 1 (HIV-1). On rare occasions, recombination can also occur between genetically distinct but distantly related retroviruses to generate a novel chimeric virus. One example is the simian immunodeficiency virus of chimpanzees (SIVcpz), which is the result of a recombination event between ancestral SIVs infecting red-capped mangabey (SIVrcp) and greater spot-nosed monkey (SIVgsn) [29, 30]. Although still controversial, SIVcpz may be the progenitor of HIV-1 [31], and it remains unclear whether this recombination had any influence on the efficiency of transmission of SIVcpz to humans. HIV-1 appears in three distinct lineages: groups M, N, and O [32–35]. Group M viruses, which are primarily responsible for the current global epidemic, can be subdivided into nine clades, A–D, F–H, J, and K, and at least 14 circulating recombinant forms (CRFs). CRFs of HIV-1 have been found in Central Africa in particular [36–38]. Although there are reports indicating that CRFs have a higher in vitro replication capacity than their parental subtypes [39], it remains unclear whether recombinants have generally advantageous properties. Recombination between HIV-1 and HIV-2 has been observed in vitro [40].
Recombination is also common among gammaretroviruses. Ecotropic FeLV-A is known to recombine with endogenous FeLV (enFeLV) env elements, yielding polytropic FeLV-B viruses [41]. Whereas FeLV-A viruses are known to have low pathogenicity, FeLV-B viruses are overrepresented in lymphosarcomas [41]. FeLV-A, which is the most transmissible form of FeLV, uses the thiamine transport protein 1 (THTR1) as cellular receptor [42]. FeLV-B has a broader in vitro host range, which includes human cells as well as a wide variety of cells derived from different animal species. FeLV-B utilizes two cell-surface receptors, PiT1 and PiT2, for infection of target cells [43, 44]. While FeLV-Bs that are able to use feline Pit2 can evolve by recombination with endogenous sequences, a subsequent point mutation during reverse transcription may be needed to generate a virus that can efficiently enter the cells using the feline Pit2 as its receptor.
Recombinants in the env gene, known as mink cell focus-inducing (MCF) viruses, play an important role between the infecting MuLV and endogenous polytropic sequences. MCF viruses have been found in most leukaemias induced by exogenous MuLV [45]. Although MCF viruses are not absolutely required for leukaemogenesis, their presence potentiates tumour development. As they infect cells via different receptors than those used by the ecotropic MuLV, MCF viruses may allow additional rounds of infection, increasing the probability of insertional activation of oncogens or inactivation of tumour suppressor genes. To investigate the interaction between endogenous and exogenous retroviruses, rat1 cells, containing a defective rat endogenous retrovirus, were infected with a defective Moloney-MuLV. Ultimately, an infectious replication-competent recombinant virus was found in this experiment [46]. These results confirm the potential for recombination between viruses from different species. The activation of PERV by an exogenous retrovirus is also a concern in the field of xenotransplantation. However, to fulfil all requirements for a recombination in this case, the infecting virus should be able to infect human cells and should contain homology regions enabling a recombination event. In addition, for retroviral recombination, copackaging of two different genomic RNAs has been proven to be a prerequisite, and therefore the packaging sites of both viruses should be compatible [47]. The 10A1 strain of MuLV provides an example of a gammaretrovirus that arose as a consequence of recombination between the exogenous 4070A MuLV and endogenous retroviral elements [48]. The recombinant 10A1 virus has an expanded host range, which results from the ability to use a novel receptor (Pit-2) in addition to the 4070A receptor Pit-1. As few as six residues in the VRA and VRB differ between the 10A1 and 4070A envelopes, and these differences have been demonstrated to account for the new host range of 10A1 [48].
Recombinant PERV-A/C
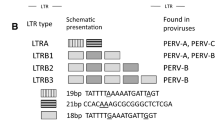
The first recombinant PERV-A/C was described by Wilson et al. [9, 10] after they infected human 293 kidney cells with supernatants from pig peripheral blood mononuclear cells (PBMCs) treated with a T cell mitogen and phorbol 12-myristate 13-acetate (PMA). At that time it was unclear whether the recombination happened in the pig or in the human cells. The breakpoints were located in the env gene (Fig. 2). LTR, gag, pol and part of env originated from PERV-C; however, a domain of env including the RBD was derived from PERV-A (Fig. 2), thus allowing the virus to replicate in human cells. Extended characterisation of this virus showed an increase in titre after repeated passages on human cells [9, 10]. Further passages resulted in higher titres, associated with genetic alterations in the LTR of the virus, mainly due to a multimerisation of transcription factor NF-Y binding sites (ATTGG) [49] (Fig. 3). Similar genetic changes in the LTR have also been seen in PERV-A passaged on human cells, although the repeats containing the NF-Y binding sites found in the LTR of PERV-A are different from the repeats in the LTR of PERV-C [49, 50] (Fig. 3). Other authors, also comparing the replication rates of PERV-A and PERV-A/C reported an isoleucine-to-valine substitution at position 140 in the RBD and changes in the proline-rich region (PRR) of the envelope protein [51]. The higher replication capacity of recombinant PERV-A/C may be associated with a higher pathogenicity, as was shown in the cases of other retroviruses, for example, HIV and FeLV.
Comparison of the genetic changes in the LTR of PERV-A and PERV-A/C during passaging on human 293 cells [49]
Similar recombinant PERV-A/Cs were detected after other co-cultivations of pig PBMCs stimulated by phytohemagglutinin and PMA with human 293 cells [52, 53] (Fig. 2). The PBMCs were derived from inbred miniature swine, and the integrated proviruses represented recombinants, all containing the receptor-binding site of PERV-A. Such recombinants were not detected in the germline of the animals.
De novo integration of PERV-A/C
Recently, the existence of recombinant PERV-A/C in pigs was described, and de novo-integrated proviruses of these recombinants were found in spleen cells of miniature pigs as well as of melanoma-bearing Munich miniature pigs, but not in the germ line of these animals [15, 53–55]. Since recombinant PERV-A/Cs are able to infect human cells, and since they increase their replication competence when passaged on human cells [9, 10, 49], they represent a novel risk for xenotransplantation.
Use of PERV-C-free pigs for a safe xenotransplantation
For xenotransplantation, the risk posed by recombinant PERV-A/C generated in pigs is obvious, because it does, in some cases, integrate de novo into the genome of pig somatic cells. It would be interesting to learn whether these viruses can infect other pigs. However, the risk coming from PERV-A/C recombinant viruses for xenotransplanation can easily be eliminated by using pigs that do not contain PERV-C in their germ line, thereby preventing recombination with PERV-A. Screening for and selection of PERV-C-free animals will reduce the risk of PERV-A/C transmission to humans.
Another reason not to use PERV-C-containing pigs for breeding to generate animals suitable for donating cells and organs for xenotransplantation is based on the ability of gammaretroviruses, including PERV, to infect cells that do not harbour the specific receptor by receptor-independent infection [56]. Most gammaretroviruses contain a PHQ motif at the amino terminus of their surface envelope protein, which is important for virus infection. In contrast, PERVs lack the full PHQ motif, having only an H residue. Mutants in this position were non-infectious but were efficiently transactivated by adding to the cells a PHQ-containing surface envelope protein derived from GaLV. A requirement of this transactivation was a functional GaLV receptor on the cells. In these experiments, transactivation by GaLV surface envelope protein enabled wild-type or H mutant PERVs of all three host-range groups to efficiently infect cells from humans and rodents that lack functional PERV receptors [56]. This ability to infect cells lacking cognate receptors was previously demonstrated for other gammaretroviruses [57, 58]. Therefore, in the presence of another retroviral surface envelope protein, for example, from a human endogenous retrovirus, and its receptor, PERV-C may infect human cells despite the fact that these cells do not express a PERV-C specific receptor.
A third reason not to use PERV-C-containing pigs is the possibility that mutations in the proline-rich domain of the Env protein of PERV-C may occur and change the tropism. Mutations in the C-terminal end of the SU protein of PERV-C have been described that resulted in binding to and infection of human cells [59].
To summarise, PERVs are, like many retroviruses, including HIV-1, prone to recombination. The risk of generating high-titre PERV-A/C recombinants as well as other risks, such as receptor-independent infection or mutation in the Env protein, which may change the tropism of PERV-C towards human cells, strongly argues against using PERV-C-containing pigs for the generation of animals intended for use in xenotransplantation.
References
Sachs DH, Sykes M, Robson SC, Cooper DK (2001) Xenotransplantation. Adv Immunol 79:129–223
Tucker A, Belcher C, Moloo B, Bell J, Mazzulli T, Humar A, Hughes A, McArdle P, Talbot A (2002) The production of transgenic pigs for potential use in clinical xenotransplantation: microbiological evaluation. Xenotransplantation 9:191–202
Denner J (1998) Immunosuppression by retroviruses: implications for xenotransplantation. Ann NY Acad Sci 862:75–86
Denner J (2007) Transspecies transmissions of retroviruses: new cases. Virology 369:229–233
Le Tissier P, Stoye JP, Takeuchi Y, Patience C, Weiss RA (1997) Two sets of human-tropic pig retrovirus. Nature 389:681–682
Patience C, Switzer WM, Takeuchi Y, Griffiths DJ, Goward ME, Heneine W, Stoye JP, Weiss RA (2001) Multiple groups of novel retroviral genomes in pigs and related species. J Virol 75:2771–2775
Ericsson T, Oldmixon B, Blomberg J, Rosa M, Patience C, Andersson G (2001) Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J Virol 75:2765–2770
Mang R, Maas J, Chen X, Goudsmit J, van der Kuyl AC (2001) Identification of novel type C porcine endogenous retrovirus: evidence that copy number of endogenous retrovirus increases during host inbreeding. J Gen Virol 82:1829–1834
Wilson CA, Wong S, Muller J, Davidson CE, Rose TM, Burd P (1998) Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol 72:3082–3087
Wilson CA, Wong S, Van Brocklin M, Federspiel MJ (2000) Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol 74:49–56
Martin U, Kiessig V, Blusch JH, Haverich A, Von der Helm K, Herden T, Steinhoff G (1998) Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692–694
Tacke SJ, Specke V, Denner J (2003) Differences in release and determination of subtype of porcine endogenous retroviruses produced by stimulated normal pig blood cells. Intervirology 46:17–24
Tacke SJ, Kurth R, Denner J (2000) Porcine endogenous retroviruses inhibit human immune cell function: risk for xenotransplantation? Virology 268:87–93
McIntyre MC, Kannan B, Solano-Aguilar GI, Wilson CA, Bloom ET (2003) Detection of porcine endogenous retrovirus in cultures of freshly isolated porcine bone marrow cells. Xenotransplantation 10:337–342
Dieckhoff B, Puhlmann J, Büscher K, Hafner-Marx A, Herbach N, Bannert N, Büttner M, Wanke R, Kurth R, Denner J (2007) Expression of porcine endogenous retroviruses PERVs in melanomas of Munich miniature swine MMS troll. Vet Microbiol 123:53–68
Suzuka I, Shimizu N, Sekiguchi K, Hoshino H, Kodama M, Shimotohno K (1986) Molecular cloning of unintegrated closed circular DNA of porcine retrovirus. FEBS Lett 198:339–343
Moennig V, Frank H, Hunsmann G, Ohms P, Schwarz H, Schäfer W (1974) C-type particles produced by a permanent cell line from a leukemic pig. II. Physical, chemical and serological characterization of the particles. Virology 57:179–188
Frazier ME (1985) Evidence for retrovirus in miniature swine with radiation-induced leukemia or metaplasia. Arch Virol 83:83–97
Patience C, Takeuchi Y, Weiss WA (1997) Infection of human cells by an endogenous retrovirus of pigs. Nat Med 3:282–286
Martin U, Winkler ME, Id M, Radeke H, Arseniev L, Takeuchi Y, Simon AR, Patience C, Haverich A, Steinhoff G (2000) Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138–142
Specke V, Rubant S, Denner J (2001) Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 285:177–180
Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP (1998) Infection of human cells by an endogenous retrovirus of pigs. J Virol 72:9986–9991
Specke V, Boller K, Schwendemann J, Denner J (2001) Porcine endogenous retroviruses (PERVs): In vitro host range and attempts to establish small animal models. J Gen Virol 82:837–844
Specke V, Schuurman HJ, Plesker R, Coulibaly C, Özel M, Langford G, Kurth R, Denner J (2002) Virus safety in xenotransplantation: preliminary data from first small animal and non-human primate in vivo experiments. Transpl Immunol 9:281–288
Specke V, Plesker R, Coulibaly C, Boller K, Denner J (2002) Productive infection of a mink cell line with porcine endogenous retroviruses (PERVs) and lack of transmission to minks in vivo. Arch Virol 147:305–319
Blusch JH, Patience C, Takeuchi Y, Templin C, Roos C, Von Der Helm K, Steinhoff G, Martin U (2000) Infection of nonhuman primate cells by pig endogenous retrovirus. J Virol 74:7687–7690
Ericsson TA, Takeuchi Y, Templin C, Quinn G, Farhadian SF, Wood JC, Oldmixon BA, Suling KM, Ishii JK, Kitagawa Y, Miyazawa T, Salomon DR, Weiss RA, Patience C (2003) Identification of receptors for pig endogenous retrovirus. Proc Natl Acad Sci 100:6759–6764
Mattiuzzo G, Matouskova M, Takeuchi Y (2007) Differential resistance to cell entry by porcine endogenous retrovirus subgroup A in rodent species. Retrovirology 4(1):93
Valérie C, Salemi M, Pourrut X, Mpoudi-Ngole E, Abela B, Auzel P, Bibollet-Ruche F, Hahn B, Vandamme AM, Delaporte E, Peeters M (2002) Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J Virol 76:8298–8309
Paraskevis D, Lemey P, Salemi M, Suchard M, Van de Peer Y, Vandamme AM (2003) Analysis of the evolutionary relationships of HIV-1 and SIVcpz sequences using bayesian inference: implications for the origin of HIV-1. Mol Biol Evol 20:1986–1996
Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BN (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441
De Leys R, Vanderborght B, van den Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems B (1990) Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of west-central Africa origin. J Virol 64:1207–1216
Gürtler LG, Hauser PH, Eberle J, von Brunn A, Knapp S, Zekeng L, Tsaque JM, Kaptue L (1994) A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol 68:1581–1585
Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Georges-Courbot MC, Barré-Sinoussi F, Brun-Vézinet F (1998) Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med 4:1032–1037
Charneau P, Borman AM, Quillent C, Guetard D, Chamaret S, Cohen J, Rémy G, Montagnier L, Clavel F (1994) Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology 205:247–253
Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Sema H, Tshimanga K, Bongo B, Delaporte E (2000) Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol 74:10498–10507
Murphy E, Korber B, Georges-Courbot MC, You B, Pinter A, Cook D, Kieny MP, Georges A, Mathiot C, Barré-Sinoussi F (1993) Diversity of V3 region sequences of human immunodeficiency viruses type 1 from the central African Republic. AIDS Res Hum Retroviruses 9:997–1006
Yang C, Dash B, Hanna SL, Frances HS, Nzilambi N, Colebunders RC, St Louis M, Quinn TC, Folks TM, Lal RB (2001) Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses 17:361–36
Konings FA, Burda ST, Urbanski MM, Zhong P, Nadas A, Nyambi PN (2006) Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes A and G. J Med Virol 78(5):523–534
Motomura K, Chen J, Hu WS (2008) Genetic recombination between human immunodeficiency virus type 1 (HIV-1) and HIV-2, two distinct human lentiviruses. J Virol 82(4):1923–1933 [Epub 2007 Dec 5]
Roy-Burman P (1995) Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes 11(2–3):147–161
Mendoza R, Anderson MM, Overbaugh J (2006) A putative thiamine transport protein is a receptor for feline leukemia virus subgroup A. J Virol 80:3378–3385
Anderson MM, Lauring AS, Robertson S, Dirks C, Overbaugh J (2001) Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J Virol 75:10563–10572
Takeuchi Y, Vile RG, Simpson G, O’Hara B, Collins MK, Weiss RA (1992) Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol 66(2):1219–1222
Fan H (1997) Leukemogenesis by Moloney murine leukemia virus: a Multistep process. Trends Microbiol 5(2):74–82
Villanueva RA, Campbell S, Roth MJ (2003) Molecular analysis of a recombinant M- MuLV/RaLV retrovirus. Virology 315(1):195–208
Hu WS, Temin HM (1990) Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA 87(4):1556–1560
Han JY, Cannon PM, Lai KM, Zhao Y, Eiden MV, Anderson WF (1997) Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J Virol 71:8103–8108
Denner J, Specke V, Thiesen U, Karlas A, Kurth R (2003) Genetic alterations of the long terminal repeat of an ecotropic porcine endogenous retrovirus PERV during passage in human cells. Virology 314:125–133
Scheef G, Fischer N, Krach U, Tönjes R (2001) The number of a U3 repeat box acting as an enhancer in long terminal repeats of polytropic replication-competent porcine endogenous retroviruses dynamically fluctuates during serial virus passages in human cells. J Virol 75:6933–6940
Harrison I, Takeuchi Y, Bartosch B, Stoye JP (2004) Determinants of high titer in recombinant porcine endogenous retroviruses. J Virol 78:13871–13879
Oldmixon BA, Woods JC, Ericsson TA, Wilson CA, White-Scharf ME, Andersson G, Greenstein JL, Schuurmann HJ, Patience C (2002) Porcine endogenous retrovirus transmission characteristics of an inbred herd of miniature swine. J Virol 76:3045–3048
Wood JC, Quinn G, Suling KM, Oldmixon BA, Van Tine BA, Cina R, Arn S, Huanq CA, Scobie L, Onions DE, Sachs DH, Schuurmann HJ, Fishman JA, Patience C (2004) Identification of exogenous forms of human-tropic porcine endogenous retrovirus in miniature swine. J Virol 78:2494–2501
Bartosch B, Stefanidis D, Myers R, Weiss R, Patience C, Takeuchi Y (2004) Evidence and consequence of porcine endogenous retrovirus recombination. J Virol 78:13880–13890
Martin SI, Wilkinson R, Fishman JA (2006) Genomic presence of recombinant porcine endogenous retrovirus in transmitting miniature swine. Virol J 3:91–96
Lavillette D, Kabat D (2004) Porcine endogenous retroviruses infect cells lacking cognate receptors by an alternative pathway: implications for retrovirus evolution and xenotransplantation. J Virol 78(16):8868–8877
Barnett AL, Wensel WL, Li W, Fass D, Cunningham JM (2003) Structure and mechanism of a coreceptor for infection by a pathogenic feline retrovirus. J Virol 77:2717–2729
Wensel DL, Li W, Cunningham JM JM (2003) A virus-virus interaction circumvents the virus receptor requirement for infection by pathogenic retroviruses. J Virol 77:3460–3469
Gemeniano M, Mpanju O, Salomon DR, Eiden MV, Wilson CA (2006) The infectivity and host range of the ecotropic porcine endogenous retrovirus, PERV-C, is modulated by residues in the C-terminal region of its surface envelope protein. Virology 346:108–117
Perryman S, Nishio J, Chesebro B (1991) Complete nucleotide sequence of Friend murine leukemia virus, strain FB29. Nucleic Acids Res 19:6950
Donahue PR, Hoover EA, Beltz GA, Riedel N, Hirsch VM, Overbaugh J, Mullins JI (1988) Strong sequence conservation among horizontally transmissible minimally pathogenic feline leukemia viruses. J Virol 62:722–731
Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J (2006) Transspecies transmission of the endogenous koala retrovirus. J Virol 80:5651–5654
Delassus S, Sonigo P, Wain-Hobson S (1989) Genetic organization of gibbon ape leukemia virus. Virology 173:205–213
Acknowledgments
The author thanks Dr. M. Eschricht, and D. Jobst for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Denner, J. Recombinant porcine endogenous retroviruses (PERV-A/C): a new risk for xenotransplantation?. Arch Virol 153, 1421–1426 (2008). https://doi.org/10.1007/s00705-008-0141-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-008-0141-7