Abstract
A false branching cyanobacterium (strain 1F-PS) isolated from a fresh water body of Bilaspur (Chhattisgarh, India) is described here as a new species of the polyphyletic genus Scytonema. Morphological, ecological, molecular and phylogenetic evidence validated the strain as a new species. Observations of the filaments in different phases of growth, different levels of microscopic studies, the presence of a textured thin sheath throughout the length of the trichome, differences in the shape and dimensions of the vegetative cells and heterocytes and ecological attributes show that the strain differed from the rest of the closely related species. Sequencing of the 16S rRNA gene resulted in 99% similarity with Scytonema bilaspurensis and 97.07% sequence similarity with Scytonema hofmannii PCC 7110, while rbcl and psbA sequences showed 99 and 97% similarities with S. bilaspurensis, respectively. Phylogenetic assessments indicated a large phylogenetic distance and separate clustering of the strain 1F-PS for all the molecular markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacterial taxonomy is a challenging field of study that is in a phase of revolution, with increasing description of new taxa using a combination of morphological, ecological and molecular data. The inherent problems of culturing, variations among the natural and laboratory grown cultures and an extremely intricate morphology, are all challenges that have made the study of cyanobacteria interesting but very complex. Taxonomic studies in the last decades (Anagnostidis and Komárek 1985, 1988, 1990; Komárek and Anagnostidis 1986, 1989; Büdel and Kauff 2012; Komárek et al. 2014) have indeed helped in solving the puzzling relationships to some level. The recent Süßwasserflora series on the cyanoprokaryotes (cyanobacteria) has recommended accommodating both morphological and genotypic data in congruence, along with more revisionary work (Komárek 2013).
The genus Scytonema is typically characterized by filaments irregularly coiled, or in fascicles, creeping or erect, which may be free, in colonies, or have tendency to form clusters, but very rarely forming layers (biofilms or strata) on the substrate. Usually the filaments have an envelope with firm colorless or maybe colored sheath. Some other features of this genus include the appearance of filaments or thallus usually in the form of clusters, or prostrate with entangled filaments and cells mostly green, olive green, blue green or yellowish (Komárek 2013). The type species is Scytonema hofmannii, and till date about 320 species have been described in this genus. Komárek et al. (2014) have placed the genus Scytonema in the family Scytonemataceae of the order Nostocales. As per the recent Süßwasserflora series, this group has been demarked as a species-rich group of false branching cyanobacteria. Previous studies (Fiore et al. 2007; Aguiar et al. 2008; Sant’anna et al. 2011; Vaccarino and Johansen 2011, 2012; Becerra-Absalón et al. 2013; Komárek 2013) have proved that the genera Scytonema and Brasilonema form a monophyletic group, based on an analysis of 16S rRNA data.
The present work, based on the freshwater strain 1F-PS, has been a challenge because of the huge diversity in the genus Scytonema and the close taxonomic affinity of this strain with a recently described strain, i.e., S. bilaspurensis (Singh et al. 2016). In spite of their closeness in habitat and ecology, comprehensive studies made it clear that the 1F-PS strain is different from S. bilaspurensis.
Materials and methods
Sampling
The sampling was performed in the Bilaspur district of Chhattisgarh State, India. Water samples were collected from a freshwater body and assessed for crucial physicochemical characteristics such as temperature, pH, total dissolved oxygen, salinity, total dissolved solids and conductivity. Immediately after collection, samples were studied through microscopy in order to assess their overall level of purity and the presence/absence of other cyanobacteria. A thorough purification of the cultures was performed using 1.0% agar plates alternating with suspension sub-culturing, repeating this process four times in order to establish the proper axenicity of the culture. The strain was grown in 150-ml basal medium (BG 11o medium) (Rippka et al. 1979). The pH of the medium was adjusted to 7.2, and the culture was maintained in culture room under an illumination of approximately 50–55 µEm−2s−1 with a photoperiod of 14/10 h light/dark cycle at 28 ± 2 °C.
Phenotypic analysis
Microscopy of the strain in different stages of growth was performed using a Nikon YS100 microscope (Nikon, Japan), and microphotographs of the cyanobacteria were taken using an Olympus BX53 microscope (Olympus Corporation, Japan) fitted with a ProgRes C5 camera (Jenoptik, USA). For better resolution and taxonomic clarity, the shape of apical cells, shape, size, orientation and other miscellaneous features of vegetative filaments, vegetative cells and heterocytes were visualized and observed. Fifty to hundred measurements were taken for each parameter. The pattern of false branching was also examined.
Molecular and phylogenetic analysis
A 10- to 15-day-old culture was used for DNA isolation and subsequent amplification using the Himedia Ultrasensitive Spin Purification Kit (MB505) with the initial lysis using the AL solution being done for 60 min, and the final lysis with the solution C1 being performed for 30 min. The amplification of the 16S rRNA and psbA genes was performed as in Singh et al. (2015b) while the rbcl gene was amplified following Singh et al. (2015a).
In the case of the 16S rRNA gene, a clean 1357 bp sequence was obtained and the similarity search was performed using the ‘Identify’ option of the EzTaxon database (http://www.eztaxon.org, Kim et al. 2012) with validated cyanobacterial strains. Similarity search was also conducted using the NCBI web services with the blast tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and retrieving only good quality and validated sequences. The rbcl and the psbA gene sequences were annotated for the coding regions using the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) and the Expasy Proteomics Server (http://expasy.org). All the sequences were submitted to the NCBI database using the sequence submission tool BankIt. Multiple sequence alignment of the sequences was performed using CLUSTAL X version 1.81 (Thompson et al. 1997). This was followed by manual alignment using DAMBE (Xia 2013). All the phylogenetic analyses were done using the software MEGA version-5 (Tamura et al. 2011). For the reconstruction of the 16S rRNA phylogeny, the model of nucleotide substitution with the lowest Bayesian Information Criterion (BIC) score, the Tamura–Nei model (Tamura and Nei 1993) (T93; BIC score of 55109.942), was selected using the MEGA 5 program. In the case of the rbcl gene, the evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter (K2) model (Kimura 1980) with a BIC score of 23,246.646. The psbA gene was analyzed by using the Maximum Likelihood method based on the Kimura 2-parameter model (K2 + I) (Kimura 1980) with a BIC score of 5294.106. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 56.3397% sites). Bootstrap values were estimated based on 1000 replications (Felsenstein 1985). The robustness of our phylogenetic findings and confirmation of the tree topology was achieved by using three different approaches for tree building: neighbor joining (NJ), maximum likelihood (ML) and maximum parsimony (MP) (Fitch 1971; Felsenstein 1981; Saitou and Nei 1987; Tamura et al. 2011).
Results
Morphological analysis
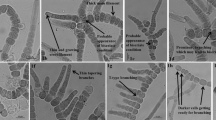
The various morphological features of the strain 1F-PS were studied carefully (Fig. 1), and comparative assessment was made with the closely related strain Scytonema hofmannii PCC 7110 and other species of the genus Scytonema. Morphological comparisons were also made with some of the other closely related genera like Petalonema and Brasilonema. Care was taken to make comparative assessments with S. bilaspurensis, which our group published recently, so that these closely related species from almost the same locality could be properly distinguished.
a Trichome showing constricted individual cells. b Filament with the tapering and curved apex. c Constricted individual cells with tapering terminal cells along with individual cells being longer than wide. d Filament with intercalary heterocyte. e Sheath breakage sites and hormogonia. f Thin filament around the sheath. g Development of false branching site. h Separation discs and individual cells being wider than long. i Filament having sheath around it and constricted individual cells along with individual cells being wider than long. j False branching sites; terminal cells with curved ends but no tapering and individual cells longer than wide. k, l Hormogonia and their formation. m False branching sites, separation discs and individual cells longer than wide. n Constricted individual cells and terminal cells having tapering ends. o–v, x, y False branching sites. w Intercalary heterocyte with two polar nodules
Molecular and phylogenetic analysis
The multiple alignments were used with proper care for phylogenetic interpretations (Online Resources 1–3). The analysis of the 16S rRNA gene revealed that the closest relatives of S. singhii were S. bilaspurensis with 99% similarity and S. hofmannii PCC 7110 with 97.07% sequence similarity. In case of the rbcl and the psbA genes similarity with S. bilaspurensis was 99 and 97%, respectively (Table 1).
In the 16S rRNA ML tree (Fig. 2), the strain 1F-PS was found to be most closely related to S. bilaspurensis, and sister to their clade was S. hofmannii PCC 7110. The bootstrap values were strong at all the nodes, and the tree topology for this marker was similar in all phylogenetic approaches. In the rbcl gene ML tree (Fig. 3), the position of S. singhii, S. bilaspurensis and S. hofmannii PCC 7110 was similar to the 16S rRNA ML tree. The MP and the NJ trees showed a phylogenetic similar to the ML tree. All the psbA gene analyses (ML, MP, and NJ) showed the strain 1F-PS (S. singhii), sister to S. bilaspurensis (Fig. 4).
Discussion
Cyanobacteria are enigmatic microbiological organisms. The severe confusion regarding their taxonomy and morphology has made their study both challenging and confusing. But, the current state of cyanobacterial taxonomy has certainly moved a bit ahead by incorporating genetic data along with studies of morphological architecture.
The genus Scytonema is a diverse genus comprising numerous species that exist endemically in tropical and ecologically distinct habitats, such as lateritic soils, dripping rocks and reservoirs with water vegetation. The genus was summarized by Geitler (1932), Desikachary (1959), Starmach (1966), Bourrelly (1970), and Komárek and Anagnostidis (1998); however, modern and recent revisions based on the polyphasic approach have indicated that this genus is much more heterogeneous and evidently polyphyletic in nature (Komárek 2013). Its heterogeneity and polyphyly has been demonstrated by molecular data (Boyer et al. 2002; Berrendero et al. 2008; Zapomělová et al. 2011), and thus, the genus has been divided into other generic units (Bohunická et al. 2012). Scytonema sensu stricto is based on the type species S. hofmannii C. Agardh ex Bornet et Flahault (1888), which is characterized by cylindrical trichomes along its whole length, more or less quadratic cells in the main trichomes, and also relatively narrow sheaths (Anagnostidis and Komárek 1988).
In this study, the strain 1F-PS showed considerable differences from the closely related strains S. bilaspurensis and S. hofmannii PCC 7110, in terms of morphology and DNA sequence data. The application of various methods like phenotypic characterization through comparative assessments and phylogenetic analyses of 16S rRNA, rbcl and psbA genetic regions proved that the strain 1F-PS was different from the other closely related species of Scytonema.
The phenotypic characterization was very conclusive in differentiating the strain 1F-PS from the closely related taxa. For the comprehensive morphological assessment, we compared the morphology of the strain 1F-PS with S. bilaspurensis (Singh et al. 2016), S. hofmannii Agardh ex Bornet et Flahault 1887, S. stuposum [Kützing] Bornet ex Bornet et Flahault 1887, Brasilonema Fiore et al. 2007, Scytonema sect. Myochrotes Bornet et Flahault 1887 and Petalonema Berkeley ex Correns 1889. The presence of a distinctly textured thin sheath, which was visible throughout the length of the trichome, was an important distinguishing feature which differentiated the strain 1F-PS from the closely related species S. hofmannii PCC 7110. It was also evident from the morphology and shape of the cells that the rectangular appearance of the cells with width usually greater than length distinguished the 1F-PS strain from S. hofmannii PCC 7110. The prominent constrictions between cells indicated that the strain 1F-PS differed significantly from S. hofmannii PCC 7110. The shape of the heterocytes was also different from what was documented by Komárek (2013) for S. hofmannii, and the presence of varying shapes of heterocytes within the 1F-PS strain was in fact an interesting feature of the strain 1F-PS. Convincing differences were also found in the size of the vegetative cells and the heterocytes which further validated our hypothesis that the strain 1F-PS was an unknown member of the genus Scytonema. Due to similarities in ecology and habitats, we documented the morphological differences of the 1F-PS strain with those of the most closely related strain S. bilaspurensis and our assessment indicates that the strains are different in terms of morphology. The strain 1F-PS was found to be much more diverse and different in the shape of the vegetative cells as compared to those of S. bilaspurensis. Also, the size of the vegetative cells and the heterocytes was also different. Thus, our comparative morphological analyses showed that there are differences between the strain 1F-PS, S. bilaspurensis and S. hofmanni PCC 7110. In addition, strain 1F-PS and S. hofmanni PCC 7110 were different in the type locality, and the presence of a non-woolly thallus in the strain 1F-PS also indicated ecological differences.
Morphological comparisons were also made with the closely related genera Petalonema and Brasilonema. It is important to note that Petalonema has not been included in a molecular phylogenetic study and that the genus is characterized morphologically by having very wide, lamellated sheaths, which are funnel-like divergent at the ends and whose sheaths are very thick and usually several times wider than the trichomes. Brasilonema presents sporadic false branching and cellular contents with dark green, grayish, reddish or even violet colors (though not always).
The close similarities of strain 1F-PS with S. bilaspurensis and S. hofmannii satisfied one of the primary criteria for species identity indicated by Strackerbrandt and Ebers (2006), who suggested a threshold limit for species identity of 98.7–99%. The polyphyly of Scytonema was noticed prominently in the 16S rRNA tree validating earlier reports of Komárek et al. (2013, 2014). According to our molecular results, the strain 1F-PS showed a close pairwise similarity of 99% with S. bilaspurensis and 94% with the strain S. hofmannii PCC 7110. The phylogenetic placement of the 1F-PS strain in the rbcl tree clearly indicated that it is different from the strains S. bilaspurensis and S. hofmannii PCC 7110. The psbA gene sequencing data indicated a 97% similarity with S. bilaspurensis, and the phylogenetic analysis showed that the 1F-PS strain and S. bilaspurensis differed in branch length.
Overall, the significant morphological, ecological and genetic differences support the recognition of the 1F-PS strain as a novel species of the genus Scytonema.
Taxonomic treatment
Scytonema singhii Singh, sp. nov.—HOLOTYPE:
India, Chhattisgarh, Bilaspur, 22.09°N, 82.15°E, Jan 2015, S.S. Singh and R.A. Minz (actively growing culture was deposited (as Scytonema sp.) at the Microbial Culture Collection (MCC), National Centre for Cell Science (NCCS), Pune, India; cryopreserved culture is maintained at the MCC (Accession Number MCC 2874)) (Fig. 1).
Etymology:
Singhii -in honor of Late. Prof. R.N. Singh, one of the India’s finest workers who studied cyanobacteria and was widely respected as one of the stalwarts of the Benares school of algology.
Description:
Blue green filaments; sometimes appearing slightly blackish green; false branching prominent; false branching appears solitary; sheath thin, colorless and narrow but perfectly distinct in both texture and color; sometimes may also take a darker shade or brownish color in older filaments; sheath present throughout the length of trichome with constrictions at the cross walls; vegetative cells ranged from 3.56 to 6.82 µm in length and 5.57–6.93 µm in width; vegetative cells are differently shaped with some being longer than wide, some cells with tapering terminal cells, some being wider than their length and some cells having curved ends but no prominent tapering; curved ends of the terminal cells of a trichome both in the main filaments and the false branches; heterocytes usually solitary; shape varying from square to cylindrical; heterocytes never have rounded ends; ranging from 4.77 to 13.05 µm in length and 5.19–6.86 µm in width.
Diagnosis:
Differing from other closely related Scytonema species in having a thin and narrow but distinctly textured sheath visible throughout the length of the trichome (Fig. 1b–f, h–j, l), individual cells prominently constricted at the cross walls (Fig. 1j). The new species differs significantly from S. bilaspurensis in having cells which may be differently shaped with some being longer than wide (Fig. 1c, j, m), some cells with tapering apical cells (Fig. 1b, c, n), some being wider than long (Fig. 1i, o), and some cells having curved ends but no prominent tapering (Fig. 1j), solitary heterocytes with varying shapes from square to cylindrical. S. singhii also differs from S. bilaspurensis in the size of the vegetative cells and the heterocytes. Genetically, it has close affinity, based on 16S rRNA, rbcl and psbA genes, with S. bilaspurensis and S. hofmannii PCC 7110, but the phylogenetic analyses indicate that S. singhii is different from those two species.
Habitat:
Fresh water body; pH 8.2; temperature 32 °C; salinity 4 g/L; total dissolved solids 100 mg/L; dissolved oxygen 70 ppm; conductivity 84.81 µS/cm.
Distribution area:
The 1F-PS strain is reported here for the first time in India, but is expected to occur in tropical and subtropical areas worldwide.
References
Aguiar R, Fiore MF, Franco MW, Ventrella MC, Lorenzi AS, Vanetti CA, Alfenas AC (2008) A novel epiphytic cyanobacterial species from the genus Brasilonema causing damage to Eucalyptus leaves. J Phycol 44:1322–1334
Anagnostidis K, Komárek J (1985) Modern approach to the classification system of the cyanophytes 1. Introduction. Algol Stud 38:291–302
Anagnostidis K, Komárek J (1988) Modern approach to the classification system of the cyanophytes 3. Oscillatoriales. Algol Stud 50:327–472
Anagnostidis K, Komárek J (1990) Modern approach to the classification system of the cyanophytes 5. Stigonematales. Algol Stud 86:1–74
Becerra-Absalón I, Rodarte B, Osorio K, Alba-Lois L, Segal-Kischinevzky C, Montejano G (2013) A new species of Brasilonema (Scytonemataceae, Cyanoprokaryota) from Tolantongo, Hidalgo, Central Mexico. Fottea 13:25–38
Berrendero E, Perona E, Mateo P (2008) Genetic and morphological characterization of Rivularia and Calothrix (Nostocales, Cyanobacteria) from running water. Int J Syst Evol Microbiol 58:447–460
Bohunická M, Mareš J, Komárek J (2012) Comparison of the molecular and morphological diversity within the family Scytonemataceae (Nostocales, Cyanobacteria). In: Anonymous, Abstract book of 14 international symposium on phototrophic prokaryotes (ISPP 2012), Porto, Portugal
Bornet É, Flahault C (1887) Revision des Nostocacées hétérocystées contenues dans les principaux herbiers de France (Troisième fragment). Ann Sci Nat Bot 5:51–129
Bornet É, Flahault C (1888) Revision des Nostocacées hétérocystées contenues dans les principaux herbiers de France (quatrième et dernier fragment). Ann Sci Nat Bot 7:177–262
Bourrelly P (1970) Les algues d’eau douce III. N. Boubée & Cie, Paris
Boyer SL, Johansen JR, Flechtner VR, Howard GL (2002) Phylogeny and genetic variance in terrestrial Microcoleus (Cyanophyceae) species based on sequence analysis of the 16S rRNA gene and associated 16S–23S ITS region. J Phycol 38:1222–1235
Büdel B, Kauff F (2012) Blue-green algae. In: Frey W (ed) Syllabus of plant families, Engler’s syllabus der Pflanzenfamilien, part VI. Borntraeger, Stuttgart, pp 5–39
Correns C (1889) Über Dickenwachstum durch Intussusception bei einiger Algenmembranen. Flora 72:1–298
Desikachary TV (1959) Cyanophyta. ICAR Monographs on Algae, New Delhi
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Molec Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fiore MF, Sant’Anna CL, Azevedo MTP, Komárek J, Kaštovský J, Sulek J, Lorenzi AS (2007) The cyanobacterial genus Brasilonema—molecular and phenotype evaluation. J Phycol 43:789–798
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool 20:406–416
Geitler L (1932) Cyanophyceae. In: Kolkwitz R (ed) Rabenhorst’s Kryptogamenflora von Deutschland. Österreich und der Schweiz 14. Algen. Akademische Verlagsgesellschaaft m. b. H., Leipzig, pp 1–1196
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Molec Evol 16:111–120
Komárek J (2013) Cyanoprokaryota. 3. Heterocytous genera. In: Büdel B, Gärtner G, Krienitz L, Schagerl M (eds) Süswasserflora von Mitteleuropa/freshwater flora of Central Europe 19/3. Springer Spektrum, Berlin, pp 1–1130
Komárek J, Anagnostidis K (1986) Modern approach to the classification system of the cyanophytes 2. Chroococcales. Algol Stud 43:157–226
Komárek J, Anagnostidis K (1989) Modern approach to the classification system of the cyanophytes 4. Nostocales. Algol Stud 56:247–345
Komárek J, Anagnostidis K (1998) Cyanoprokaryota 1. Chroococcales. In: Ettl H, Gärtner G, Heynig H, Mollenhauer D (eds) Süsswasserflora von Mitteleuropa 19/1. Gustav Fischer, Jena, pp 1–548
Komárek J, Kaštovský J, Mareš J, Johansen JR (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 86:295–335
Rippka R, Deruelles J, Waterbury JB, Herdman MR, Stanier RY (1979) Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. J Gen Microbiol 111:1–61
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molec Biol Evol 4:406–425
Sant’anna CL, Azevedo MTP, Fiore MF, Lorenzi AS, Kaštovský J, Komárek J (2011) Subgeneric diversity of Brasilonema (Cyanobacteria, Scytonemataceae). Revista Brasil Bot 34:51–62
Singh P, Fatma A, Mishra AK (2015a) Molecular phylogeny and evogenomics of heterocystous cyanobacteria using rbcl gene sequence data. Ann Microbiol 65:799–807
Singh P, Singh SS, Aboal M, Mishra AK (2015b) Decoding cyanobacterial phylogeny and molecular evolution using an evonumeric approach. Protoplasma 252:519–535
Singh P, Minj RA, Kunui K, Shaikh ZM, Suradkar A, Shouche YS, Mishra AK, Singh SS (2016) A new species of Scytonema isolated from Bilaspur, Chhattisgarh, India. J Syst Evol 5:519–527. doi:10.1111/jse.12202
Starmach K (ed) (1966) Flora slodkowodna Polski 2. Cyanophyta-sinice, Glaukophyta-glaukofity. Panstwowe Wydawnictwo Naukowe, Warszawa
Strackerbrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 33:152–155
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molec Biol Evol 10:512–526
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molec Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vaccarino MA, Johansen JR (2011) Scytonematopsis contorta sp. nov. (Nostocales), a new species from the Hawaiian Islands. Fottea 11:149–161
Vaccarino MA, Johansen JR (2012) Brasilonema angustatum sp. nov. (Nostocales), a new filamentous cyanobacterial species from the Hawaiian Islands. J Phycol 8:1178–1186
Xia X (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Molec Biol Evol 30:1720–1728
Zapomělová E, Hrouzek P, Řezanka T, Jezberová J, Řeháková K, Hisem DJR, Komárková J (2011) Polyphasic characterization of Dolichospermum spp. and Sphaerospermopsis spp. (Nostocales, Cyanobacteria): morphology, 16S rRNA gene sequences and fatty acid and secondary metabolite profiles. J Phycol 47:1152–1163
Acknowledgements
We are thankful to the Council of Scientific and Industrial Research, New Delhi, India for financial support in the form of project (Ref. No. 38(1333)/12/EMR(II). The Head Department of Botany, Guru Ghasidas Vishwavidyalaya, Bilaspur, Chhattisgarh, India is gratefully acknowledged for providing laboratory facilities. We thank the Director NCCS for facilities and encouragement. Authors from NCCS are thankful to the Department of Biotechnology (DBT; Grant no. BT/PR/0054/NDB/52/94/2007), the Government of India, under the project ‘Establishment of Microbial Culture Collection.’
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Ricarda Riina.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Information on Electronic Supplementary Material
Information on Electronic Supplementary Material
Online Resource 1 . 16S rRNA alignment file with the strain 1F-PS and all the considered taxa.
Online Resource 2 . rbcl alignment file with the strain 1F-PS and all the considered taxa.
Online Resource 3 . psbA alignment file with the strain 1F-PS and all the considered taxa.
Rights and permissions
About this article
Cite this article
Singh, P., Minz, R.A., Kunui, K. et al. A new species of Scytonema isolated from Bilaspur, Chhattisgarh, India using the polyphasic approach. Plant Syst Evol 303, 249–258 (2017). https://doi.org/10.1007/s00606-016-1370-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-016-1370-y