Abstract
A sensing platform is presented for the determination of alkaline phosphatase (ALP) activity based on the cooperation of DNAzyme-Au spherical nucleic acid nanoprobe with the graphene-oxide–loaded hybridization chain reaction (HCR/GO) system to achieve good detection sensitivity and specificity. This assay takes advantage of the strong affinity of pyrophosphate (PPi) to Cu2+ ions and the fact that ALP can hydrolyze pyrophosphate (PPi) to release free Cu2+ ions. In the presence of ALP, the released Cu2+ can promote the Cu2+-dependent DNAzyme to cleave the substrate that generates a shorter DNA fragment, which is responsible for further triggering the HCR/GO system to form a long fluorescence dsDNA and thereby giving an amplified fluorescence signal. Linear calibration range was obtained from 0.2 to 10 U L−1, and the limit of detection (LOD) is about 0.14 U L−1. The feasibility of the proposed method was validated by spiking ALP standards in bovine serum. The recovery ranged from 97.2 to 104.6%, and a coefficient of variation (CV) of less than 8% (n = 3) was obtained. This assay strategy was also applied to evaluate the ALP inhibitor efficiency, which indicates that the assay has potential for drug screening.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As a critical mediator of biological systems, alkaline phosphatase (ALP) actively participates in the physiological processes of the body, which plays an integral role in metabolism, signal transduction, molecule transportation, and genetic expression information [1,2,3,4]. The normal level of ALP activity in human serum is in a range of 40 to 150 U L−1 [5]. Abnormal ALP activity has been universally recognized as a marker for various diseases such as bone disease, liver dysfunction, breast, and prostate cancer [4,5,6,7]. Of note, disease spectrums associated with abnormal ALP activity are still developing, which causes a higher demand for ALP activity detection to further understand ALP’s role in physiological processes [5, 8, 9]. Therefore, developing analytical methods for measuring ALP activity with high sensitivity and precision is of considerable significance in clinical diagnoses.
To date, many strategies have been developed for ALP activity assay based on the dephosphorylation by ALP. In general, specially designed substrates such as p-nitrophenyl phosphate, 4-methylumbellyferyl phosphate, and p-aminophenyl phosphate are dephosphorylated by ALP to produce colored (p-nitrophenol), highly fluorescent (4-methylumbellyferyl), or electroactive (p-aminophenyl) products that are straightly detected by spectroscopic or electrochemical methods [6, 10, 11]. Despite being characterized by straight-forward and fast response, these methods are limited to clinical tests due to low sensitivity. Apart from the synthetic small molecules that contain phosphate groups, pyrophosphate (PPi), a natural substrate of ALP is often utilized for ALP activity assay based on the strong binding of PPi to specific metal ions (including Al3+, Zn2+, Cu2+, Fe3+, and Ce3+) and the fact that ALP can efficiently catalyze the hydrolysis of PPi to phosphate (Pi), a process that generates uncomplexed metal ions [6, 12,13,14]. According to the design, the amount of released metal ions reflects the activity of ALP and is consistent with the concentration of ALP in the samples, which is determined by the quenching or stimulating influences on various foreign fluorescent labels, such as quantum dots (QDs) [15], metal-based nanomaterials [12], and upconversion materials [16]. However, most of these methods suffer from false-positive signals as a consequence of its sensing mechanism toward metal ions [6], although they have made many achievements.
DNAzymes, which was obtained via in vitro selection, have received great significance due to their unique recognition ability toward specific metal ions and the catalytic properties [17]. Because of such high metal ion selectivity, in combination with the excellent chemical stability, synthetic accessibility, as well as ease of modification with various functional molecules, these DNAzymes have been a powerful tool in the design of sensors for many metal ions, such as Zn2+, Cu2+, UO22+, and Pb2+ [18,19,20,21]. More importantly, as ssDNA strands, DNAzymes can be further incorporated into other nucleic acid amplification techniques (such as RCA, SDA, and hybridization chain reaction (HCR)) for enhancing the detection sensitivity [21,22,23,24].
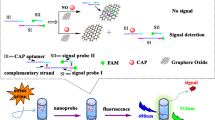
Herein, taking advantage of these excellent properties of DNAzyme, we report a sensing platform for ALP activity assaying. As shown in Scheme 1a, a Cu2+-dependent DNAzyme (Cu-Enz)–functionalized gold nanoparticle (DNAzyme-Au) nanoprobe was used as the recognition component for monitoring the free Cu2+ ions generated via ALP-catalyzed hydrolysis of PPi. To construct this nanoprobe, 20 nm AuNPs was chosen as a scaffold for assembling and orienting the thiol-modified substrate stands into a dense arrangement, followed by hybridizing with the Cu-Enz. Upon hydrolyzation by ALP, free Cu2+ ions are present that activate the DNAzymes to cleave the intact substrate DNA strands (Cu-Sub), which results in the disassociation of a shorter DNA fragment (named as trigger-DNA) from the AuNPs due to lack of thermal stability.
However, regarding the DNAzyme-Au nanoprobe, the fluorescence signal was switched on in a one-to-one form with the ALP catalytic event, which would limit the detection sensitivity. Therefore, a graphene-oxide–loaded hybridization chain reaction (HCR/GO) system that was initiated by the trigger DNA, was incorporated with nanoprobe for tandem signal amplification. The kinetics-controlled HCR reaction is an ideal signal amplification tool for detecting specific nucleic acids because it does not require any enzyme reactions [25]. At the same time, the graphene oxide (GO) can be used as the carrier to accumulate the HCR products that enrich the fluorescence signal [26, 27]. As a result of the amplification, the signal-to-background value (S/N) was improved more than 12-fold compared to using the single DNAzyme-Au nanoprobe. Incorporation of the HCR/CO system with the nanoprobe leads to a novel ALP activity assay platform that possesses excellent specificity and sensitivity.
Experiment section
Chemicals and instruments
Alkaline phosphatase from bovine intestinal mucosa was bought from Sigma-Aldrich. Chloroauric acid (HAuCl4·4H2O, 99.99%) was obtained from Shanghai Chemical Reagent Company (Shanghai, China). The GO was bought from XFNANO Materials Tech Co., Ltd. (Nanjing, China), which was then ultrasound according to the instruction from the manufacturer to obtain the GO nanosheets (0.5 mg mL−1). Oligonucleotides used in this work were custom synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China), and the sequences and modifications are listed in Table S1. All the other chemicals were of analytical grade and obtained from commercial sources without further purification. The aqueous solutions of all the reagents were prepared using deionized water (≥ 18 MΩ, Watsons). Transmission electron microscopy (TEM) images of AuNPs was carried out on JEM-2100 TEM (JEOL Ltd., Japan). A Hitachi U-3010 UV-vis spectrometer (Japan) was used to measure the absorption spectra. Fluorescence signal was obtained on a Hitachi F-7000 spectrometer (Hitachi, Japan).
Preparation of DNAzyme functionalized AuNPs
Twenty-nanometer AuNPs was used to construct the DNAzyme-Au nanoprobe, and the synthesis process is shown in the “Supplementary information.” AuNPs were first functionalized with thiolated Cu-Sub, followed by hybridizing with the Cu-Enz. Specifically, 10 μL of Cu-Sub (100 μM), 10 μL of TCEP (10 mM), and 1 μL of NaAc-HAc buffer (500 mM, pH 5.2) were mixed and kept for 1 h to reduce the disulfide bond. The reduced Cu-Sub was mixed with 750 μL of AuNP solution, adding 75 μL 20% Tween20, and incubated for 5 min; then adding 562 μL 5 M NaCl (2 M final concentration) to the mixture to shield the charge repulsion among the neighboring DNAs, thereby enhancing the DNA loading amounts. After the addition of NaCl, the following steps were repeated three times consisting of sonication for 2 min, followed by incubation for 30 min. Unconjugated DNA was separated from the AuNPs by centrifugation (13,000×g for 30 min), and the red precipitate was washed three times with 10 mM PBS buffer (pH 7.4) containing 0.3 M NaCl, which was then resuspended in 200 μL of the same buffer. In the subsequent hybridization process, 10 μL 100 μM Cu-Enz was added to the Cu-Sub-AuNP solution. The solution was maintained at 55 °C for 5 min and allowed to cool to room temperature for 1 h to achieve full hybridization. The product DNAzyme-Au nanoprobe was isolated by centrifugation (13,000×g, 20 min), then washed twice with 10 mM PBS buffer (200 mM NaCl, pH 7.4) and finally resuspended in this PBS for further use.
Optimize the working concentration of Cu2+ and PPi
To achieve the optimized response to ALP, the optimal working concentration of Cu2+ ions was first explored by examining the intensities of the fluorescence signals arising from the DNAzyme-Au nanoprobe. A series of concentrations of Cu2+ stock solution was added into the nanoprobe solution, which was incubated for 20 min at room temperature, and the intensities of their fluorescence signals were recorded. The optimal concentration of PPi required to block Cu2+ was determined next; the solution of PPi with different concentrations was mixed with 10 μM of Cu2+, followed by incubation for 30 min at room temperature to enable a sufficient combination of PPi and Cu2+. Then, FAM-modified nanoprobe solution (0.5 nM) was added to this mixture. Finally, the fluorescence changes were recorded after incubation for 20 min at 37 °C.
Preparation of HCR/GO system
The HCR/GO system was synthesized as previously described [26]. Five microliters of each FAM-labeled hairpin DNA (10 μM) was first annealed to form the stable hairpin structure, which was mixed with a GO solution (500 μg mL−1 of 60 μL) and incubated for 1 h at room temperature. The mixture was centrifuged for 15 min at 15,000 rpm to remove excess hairpin DNA strands. The sediments were resuspended in 100 μL of PBS buffer and stored at 4 °C before used.
Detection procedure of ALP activity
In the typical experiment for ALP activity assay, 1 μL of Cu2+ (1 mM), 6 μL of PPi (1 mM), and 10 μL of a sample containing a certain concentration of ALP were mixed in a reaction buffer (10 mM, Tris-HCl, 200 mM NaCl), making a total volume of 80 μL. This mixture was incubated for 30 min at 37 °C for the ALP catalytic reaction. Then, 20 μL DNAzyme-AuNP probe (final concentration was 0.5 nM) was added, followed by incubation for 20 min at room temperature. A solution (30 μL) of HCR/GO (300 μg mL−1) and 3 μL of EDTA (10 mM) was finally added to the above solutions, and the resulting solution was held at 37 °C for 2 h before the fluorescence spectra were recorded. In the whole analysis process, the fluorescence spectrums were collected between 500 and 600 nm using the maximal excitation wavelength at 492 nm.
ALP detection procedure in serum samples
For ALP detection in bovine serum, 10 μL bovine serum was spiked with 10 μL of solutions containing varying concentrations of ALP and then diluted with 980 μL of reaction buffer (10 mM, Tris-HCl, 200 mM NaCl) to give a total volume of 1 mL. One microliter of Cu2+ (1 mM) and 6 μL of PPi (1 mM) were mixed with aliquots (73 μL) of the diluted serum solutions (1%) containing ALP, which is then incubated at 37 °C for 30 min for the ALP catalytic reaction. The detection procedure was the same as described in the “Detection procedure of ALP activity” section.
Results and discussion
Characterization of the DNAzyme-Au Nanoprobe
Since the pioneering work on DNA-AuNP nanoconjugates by Mirkin [28], DNA-AuNP has been one of the most versatile hybrid nanomaterials for bioanalysis. Compared with those linear oligonucleotide-based sensors, the spherical nucleic acid nanoprobe that is heavily loaded with oligonucleotide has significant advantages in detecting sensitivity and specificity [29]. Thus, we utilize 20 nm AuNP as the scaffold, followed by functionalizing the AuNPs with DNAzyme and its substrate to construct the nanoprobe. The synthesis of AuNPs is shown in the “Supplementary information.” Bare AuNPs were synthesized and characterized by TEM. The TEM image (Fig. S1a) indicates that the spherical AuNPs were well prepared with an average size of 20 nm. Subsequently, the Cu-Sub strands modified with a thiol group in 3′ side were immobilized onto the AuNPs’ surface via Au–S bond and then hybridized with Cu-Enz strands. To enhance the accessibility of the Cu-Sub strand to the Cu-Enz, a 20-thymine (T) spacer was added between the thiol moiety and the enzyme strands. The UV absorption spectra of the DNA-modified AuNPs (Fig. S1b) show a slight shift compared to the typical AuNP peak at 520 nm. But the DLS-determined hydrodynamic size of AuNPs changed obviously (Fig. S1c). For fluorophore-labeled DNAzyme-AuNP nanoprobe, the 5′ end of the substrate is labeled with a FAM molecule whose fluorescence is quenched by the AuNP. The amount of hybridized DNAzyme covering the per AuNP surface was determined by a fluorescence-based method that employs a mercaptoethanol competing experiment. Fluorescence quantification assay based on the standard linear calibration curves of FAM-labeled substrate stands (Fig. S2) shows that there are approximately 140 hybridized DNAzyme strands on each AuNP. Thus, the parameters for the DNAzyme-Au nanoprobe is set at a stoichiometry of approximately 140 DNA strands per particle. Such a dense loading of DNAzymes and efficient hybridization between the DNAzyme and their substrate strand allows for maximum dynamic range.
Optimization of the concentration of Cu2+ ions
The free Cu2+ that is generated via ALP hydrolyzing of the PPi-Cu2+ complex can be regarded as an indicator to reflect the activity of ALP in samples. Therefore, the nanoprobes that can reliably monitor the concentration of Cu2+ is crucial for ALP activity assay. We thus investigated the fluorescence increment of the FAM-modified DNAzyme-AuNPs (0.5 nM particle concentration, corresponding to 70 nM substrate) in the presence of different concentrations of Cu2+ (Fig. 1a) to test the response of nanoprobe to Cu2+. As shown in Fig. 1b, the fluorescence intensity increased gradually with increasing concentration of Cu2+ and exhibited good linear correlation (R2 = 0.9923) when the Cu2+ concentration lies between 0.01 and 10 μM, suggesting that the DNAzyme-Au nanoprobe is reliable for monitoring the Cu2+ ions. While it should be noted that the fluorescence intensities decreased if the Cu2+ concentration was more than 30 μM, which is reasonable to believe that the high Cu2+ concentration would reduce the system stability. Therefore, the best performance of the DNAzyme-Au nanoprobe can be achieved at 10 μM Cu2+. We also tested the nanoprobe in response to various biologically relevant metal ions (including Mg2+, Fe3+, Ca2+, K+, Zn2+, and Mn2+) to evaluate its selectivity to Cu2+. As Fig. 1c shows, 10 μM of Cu2+ can induce about 60% cleavage efficiency of DNAzyme that is much higher than other competing metal ions, indicating that the nanoprobe has a good selectivity to Cu2+.
a Fluorescence response of DNAzyme-AuNPs at different concentrations of Cu2+ (λex = 492 nm). b The relationship of the fluorescence enhancement with the Cu2+ concentration. Inset: the linear responses of DNAzyme-Au nanoprobe to Cu2+. Error bars show the standard deviations of three experiments. c Specificity of the nanoprobe over Cu2+ and other metal ions. d Fluorescence responses of DNAzyme-AuNPs at treatment with 10 μM Cu2+ in the presence of varying concentrations of PPi (λex = 492 nm). Inset: the calibration curves of fluorescence intensity to PPi concentrations. Error bars show the standard deviations of three experiments
Optimization of the concentration of PPi
We then investigated the effect of PPi concentration to obtain the optimum working concentration of PPi for ALP assay. The amount of PPi is a crucial factor that influences the background signal and detection sensitivity. In principle, an appropriate level of PPi should be required to completely block Cu2+ that prevents the DNAzyme-Au nanoprobe from being active in the absence of ALP. Hence, the working concentration of PPi was determined by using the optimized Cu2+ level. With the increase of PPi from 0 to 60 μM, as shown in Fig. 1d, the fluorescence intensity of nanoprobe gradually decreases because PPi binds to Cu2+. The fluorescence intensity reaches the lowest value at PPi concentrations above 60 μM, implying that the Cu2+ can be completely bound when the concentration of PPi is above 60 μM. Then, 60 μM of PPi was thus used for the following experiment.
Studies of catalytic kinetic of ALP with DNAzyme-Au nanoprobes
The catalytic kinetic of ALP was studied by investigating the time-dependent fluorescence enhancements of the FAM-modified nanoprobe in the presence of different concentrations of ALP (Fig. 2). Within the reaction time of 30 min, the rate of fluorescence enhancement was increased with the increase of ALP concentration. The fluorescence spectrum of the DNAzyme-Au nanoprobe with different concentrations of ALP was recorded at 30 min (Fig. 3a), and a curve between fluorescence change ratio (F/F0–1) and ALP concentration ranging from 0.2 to 10 U L−1 is plotted in Fig. 3b. An excellent linear relationship F/F0–1 = 0.293 [ALP] + 0.580 (R2 = 0.9913) exists in this plot in the range of 0.2 to 10 U L−1. However, within the nanoprobe detection system, each DNAzyme strand is only responsible for cutting one fluorescent substrate strand and releases one fluorophore from the AuNP surface, which would offer limited sensitivity.
a Fluorescence response of DNAzyme-Au nanoprobe after reaction of 30 min in the presence of different concentrations of ALP (λex = 492 nm). b Plot of fluorescence change ratio (F/F0–1) value vs. the concentration of ALP that ranges from 0 to 40 U L−1. Inset: plot of linear region from 0.2 to 10 U L−1. Error bars show the standard deviations of three experiments
The design of amplified sensing system for ALP
Therefore, a GO-load HCR system was further proposed as a signal amplification tool to improve detection sensitivity, in which a single trigger DNA that arises from the DNAzyme cleaves its substrate strand, could trigger the self-assembly of HCR. As strong as noncovalent binding of GO with nucleobases and aromatic compounds was reported [25,26,27], the GO was used to bind FAM-labeled hairpin DNA and quenched the fluorescence of FAM. The proposed HCR/GO system was constructed by absorbing two FAM-modified hairpin sequences (H1 and H2, respectively) onto GO surface via π stacking. Once the hybridization of the trigger DNA to hairpin DNA initiates the HCR, a long dsDNA product with accumulated fluorescence signals is formed followed by a desorption from GO nanosheet due to the weaker affinity between GO nanosheet and dsDNA. Based on this design principle, the DNA structures used for constructing the DNAzyme-Au nanoprobe was designed and schematically shown in Scheme 1b. The hybridization of Cu-Enz strand to Cu-Sub strand forms a complex asymmetric manner, in which the 3′ portion of the substrate is hybridized with the DNAzyme through Watson-Crick base pairs, and the 5′ part contains 8 bases that bind to DNAzyme via the formation of a triple-helix conformation. When treating with Cu2+, the DNAzyme is activated, cleaving the substrate strand at the guanine site, generating two DNA fragments F1 and F2. The F1 is what we call the trigger DNA responsible for trigger HCR/GO, which was dissociated with arm 2 and released from the AuNP surface.
We thus verified if the protocol of the sensing platform with HCR/GO is competent for ALP assay, especially on whether the HCR/GO could keep the nanoprobe silent in the absence of ALP. A fluorescence test was conducted to evaluate the leakage phenomenon of the sensing platform by co-incubating the nanoprobe with HCR/GO. The result in Fig. 4a shows that there is no significant change in the fluorescence intensity of the system after the HCR/GO co-incubation with nanoprobe for 1 h, suggesting that the HCR/GO is inert to unactivated nanoprobe. We also performed the same experiments on the incubation of HCR/GO with PPi and Cu2+ separately. Similar results show that these individual components cannot cause interference for ALP assay. When ALP is introduced, the Cu2+ cofactor of DNAzyme was released by ALP hydrolyzing the PPi; the nanoprobe was activated to cleave the substrate strand and releases the trigger DNA, thereby initiating the HCR to recover the fluorophore that is quenched by GO. This process is demonstrated by the significantly increased fluorescence of the sensing system, which further confirmed that the sensing system is ALP activity dependent. More importantly, the enhanced fluorescent intensity accompanied by the DNAzyme-Au nanoprobe and the HCR/GO system illustrated higher sensitivity that could be obtained by the combination sensing platform.
a Fluorescence responses of control experiments under different conditions: (a) DNAzyme-Au nanoprobe+HCR/GO system (no ALP); (b) DNAzyme-Au nanoprobe+HCR/GO system (10 U L−1 ALP); (c) DNAzyme-Au nanoprobe (no ALP); (d) DNAzyme-Au nanoprobe (10 U L−1 ALP); (e) PPi+HCR/GO system; and (f) Cu2++HCR/GO system. For all experiments, the concentration of PPi and Cu2+ was 10 μM and 60 μM, respectively. b Fluorescence spectra of the AuNP-HCR/GO system with addition of different concentrations of ALP. Inset: the linear curve between the fluorescence change ratio (F/F0–1) value and ALP concentration ranging from 0.2 to 10 U L−1. Error bars show the standard deviations of three experiments
Analytical performance of the sensing system for ALP activity assay
The sensitivity and selectivity of the entire assay were systematically investigated. The activity of ALP was plotted against the fluorescence change ratio (F/F0–1), and they were fitted to a linear curve F/F0–1 = 4.230 [ALP] + 1.534 (R2 = 0.9907) with a dynamic range from 0.2 to 10 U L−1 (Fig. 4b). The detection limit is estimated to be 0.144 U L−1 based on 3σ/slope. Additionally, there is a nearly 12-fold enhancement of S/N ratio for the AuNP-HCR/GO sensing system compared with that of the DNAzyme-Au nanoprobe alone. We prepared several interference proteins, including bovine serum albumin (BSA), horseradish peroxidase (HPR), glucose oxidase (GOx), human serum albumin (HSA), and protein kinase (PKA), to evaluate the selectivity of the sensing platform. As shown in Fig. S6, these five proteins at 10 μg mL−1 concentrations did not yield obvious fluorescence signals, whereas the ALP at a concentration 200 times lower (≈ 5 ng mL−1) resulted in a large fluorescence increase, which demonstrates that the sensing platform is selective for ALP.
ALP activity assay in biological samples
We thus examined the response of the sensing system to ALP in diluted human serum (1%). However, the activity of ALP in unspiked bovine serum is almost undetectable though ALP is a component in the serum, which perhaps allows ALP to lose its activity during bovine serum storage. We further studied the recovery of this method by adding a known amount of ALP into the serum samples (Table 1). Varying concentrations of ALP was spiked into the bovine serum follow by diluting the sample to 1%, and an acceptable recovery ratio that ranges from 97.2 to 104.6% was achieved, with a coefficient of variation (CV) that is less than 8% (n = 3). These results indicate that our sensing platform could be potentially used to evaluate the ALP activity in practical biological samples.
Evaluate the ALP inhibitor efficiency
Moreover, the practicability of the sensing platform was demonstrated by its utility in screening the ALP inhibitors. Here, NaF was chosen as a model inhibitor, and its inhibition effect on the ALP was tested by using the developed method. ALP (10 U L−1) was first treated with different concentrations of NaF for 30 min, and then the activity of NaF-treated ALP was measured. In Fig. 5, a profile was obtained by plotting the system fluorescence intensity against the NaF concentration. The measured IC50 value of NaF was calculated to be about 1.1 ± 0.2 mM, which is consistent with previous reports [10, 30]. This result demonstrated that the sensing platform could be used to evaluate the inhibitor efficiency and may further be applied to screen other ALP inhibitors.
Comparison of the current method to other techniques
Also, various sensing platforms with different mechanisms have been explored to determine ALP activity. To assess the performance of ALP detection, we compared this assay with other methods. As shown in Table S1, the LOD and linear range of ALP based on colorimetric, electrochemical, and fluorescence are summarized. Most of these strategies and our method acquired a satisfying analytical performance for determining ALP levels in biological samples. Compared with other methods, our sensing platform is a DNA-based fluorescence strategy, which shows several obvious advantages, such as simplicity for design and synthesis, low cost, and excellent biocompatibility. Meanwhile, given the recognition ability of DNAzyme-Au nanoprobe and HCR/GO to act as a signal amplifier, our sensing platform may be easily adapted to be extended to detect other targets using DNA hybridization technology. Nevertheless, the operation of this sensing platform in practical applications is slightly cumbersome since the analysis process requires the participation of two components.
Conclusion
In this work, we established a novel signal amplification sensing platform for highly sensitive ALP activity detection. The sensing platform was built on the mechanism that ALP can regulate the concentration of free Cu2+ released from the PPi-Cu2+ complex. The platform contains a DNAzyme-based nanoprobe as the Cu2+ ion recognition component and an HCR/GO system as the signal amplifier. These designs together allowed a high sensitivity for ALP assay and resulted in an approximately 12-fold increase in the S/N ratio compared with the DNAzyme-Au nanoprobe alone. The practical application in biological samples of this strategy was confirmed by using a spiked bovine serum as the ALP-positive sample. This sensing platform contains the first successful integration of DNAzyme-Au nanoprobe with the HCR/GO system for the determination of ALP activity. Given the performance of high sensitivity and reliability, this sensing platform has been successfully used in evaluating the ALP inhibitor efficiency. The assay has the potential as a platform for clinical ALP assay or for screening new ALP inhibitors.
References
Siller AF, Whyte MP (2018) Alkaline phosphatase: discovery and naming of our favorite enzyme. J Bone Miner Res 33:362–364
Millán JL (2006) Alkaline phosphatases. Purinergic Signal 2:335–341
Han Y, Chen J, Li Z, Chen H, Qiu H (2020) Recent progress and prospects of alkaline phosphatase biosensor based on fluorescence strategy. Biosens Bioelectron 148:111811
Zhao WW, Xu JJ, Chen HY (2015) Photoelectrochemical bioanalysis: the state of the art. Chem Soc Rev 44:729–741
Song Z, Kwok RTK, Zhao E, He Z, Hong Y, Lam JWY, Liu B, Tang BZ (2014) A ratiometric fluorescent probe based on ESIPT and AIE processes for alkaline phosphatase activity assay and visualization in living cells. ACS Appl Mater Inter 6:17245–17254
Tang Z, Chen H, He H, Ma C (2019) Assays for alkaline phosphatase activity: progress and prospects. TrAC-Trend Anal Chem 113:32–43
Niu X, Ye K, Wang L, Lin Y, Du D (2019) A review on emerging principles and strategies for colorimetric and fluorescent detection of alkaline phosphatase activity. Anal Chim Acta 1086:29–45
Jin C, He J, Zou J et al (2019) Phosphorylated lipid-conjugated oligonucleotide selectively anchors on cell membranes with high alkaline phosphatase expression. Nat Commun 10:1–7
Zhang L, Nie JF, Wang HL, Yang JH, Wang BY, Zhang Y, Li JP (2017) Instrument-free quantitative detection of alkaline phosphatase using paper-based devices. Anal Methods 9:3375–3379
Sun J, Zhao J, Bao X, Wang Q, Yang X (2018) Alkaline phosphatase assay based on the chromogenic interaction of diethanolamine with 4-aminophenol. Anal Chem 90:6339–6345
Park KS, Lee CY, Park HG (2014) A sensitive dual colorimetric and fluorescence system for assaying the activity of alkaline phosphatase that relies on pyrophosphate inhibition of the peroxidase activity of copper ions. Analyst 139:4691–4695
Ma JL, Yin BC, Wu X, Ye BC (2016) Copper-mediated DNA-scaffolded silver nanocluster on-off switch for detection of pyrophosphate and alkaline phosphatase. Anal Chem 88:9219–9225
Kong RM, Fu T, Sun NN, Qu FL, Zhang SF, Zhang XB (2013) Pyrophosphate-regulated Zn2+-dependent DNAzyme activity: an amplified fluorescence sensing strategy for alkaline phosphatase. Biosens Bioelectron 50:351–355
Das P, Chandar NB, Chourey S, Agarwalla H, Ganguly B, Das A (2013) Role of metal ion in specific recognition of pyrophosphate ion under physiological conditions and hydrolysis of the phosphoester linkage by alkaline phosphatase. Inorg Chem 52:11034–11041
Kang W, Ding Y, Zhou H, Liao Q, Yang X, Yang Y, Jiang J, Yang M (2015) Monitoring the activity and inhibition of alkaline phosphatase via quenching and restoration of the fluorescence of carbon dots. Microchim Acta 182:1161–1167
Wang F, Zhang C, Xue Q, Li H, Xian Y (2017) Label-free upconversion nanoparticles-based fluorescent probes for sequential sensing of Cu2+, pyrophosphate and alkaline phosphatase activity. Biosens Bioelectron 95:21–26
Zhou W, Saran R, Liu J (2017) Metal sensing by DNA. Chem Rev 117:8272–8325
Yin BC, Zuo P, Huo H, Zhong X, Ye BC (2010) DNAzyme self-assembled gold nanoparticles for determination of metal ions using fluorescence anisotropy assay. Anal Biochem 401:47–52
Wu P, Hwang K, Lan T, Lu Y (2013) A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J Am Chem Soc 135:5254–5257
Tang Z, Zhang H, Ma C, Gu P, Zhang G, Wu K, Chen M, Wang K (2018) Colorimetric determination of the activity of alkaline phosphatase based on the use of Cu (II)-modulated G-quadruplex-based DNAzymes. Microchim Acta 185:109
Peng H, Newbigging AM, Wang Z, Tao J, Deng W, Le XC, Zhang H (2018) DNAzyme-mediated assays for amplified detection of nucleic acids and proteins. Anal Chem 90:190–207
Peng H, Newbigging AM, Reid MS et al (2019) Signal amplification in living cells: a review of microRNA detection and imaging. Anal Chem 92:292–308
Zhang Y, Fan J, Nie J, Le S, Zhu W, Gao D, Yang J, Zhang S, Li J (2015) Timing readout in paper device for quantitative point-of-use hemin/G-quadruplex DNAzyme-based bioassays. Biosens Bioelectron 73:13–18
Gao R, Xu L, Hao C, Xu C, Kuang H (2019) Circular polarized light activated chiral satellite nanoprobes for the imaging and analysis of multiple metal ions in living cells. Angew Chem 131:3953–3957
Hong M, Xu L, Xue Q, Li L, Tang B (2016) Fluorescence imaging of intracellular telomerase activity using enzyme-free signal amplification. Anal Chem 88:12177–12182
Si H, Sheng R, Li Q, Feng J, Li L, Tang B (2018) Highly sensitive fluorescence imaging of Zn2+ and Cu2+ in living cells with signal amplification based on functional DNA self-assembly. Anal Chem 90:8785–8792
He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C (2010) A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv Funct Mater 20:453–459
Cutler JI, Auyeung E, Mirkin CA (2012) Spherical nucleic acids. J Am Chem Soc 134:1376–1391
Yang Y, Zhong S, Wang K, Huang J (2019) Gold nanoparticle based fluorescent oligonucleotide probes for imaging and therapy in living systems. Analyst 144:1052–1072
Shen C, Li X, Rasooly A, Guo L, Zhang K, Yang M (2016) A single electrochemical biosensor for detecting the activity and inhibition of both protein kinase and alkaline phosphatase based on phosphate ions induced deposition of redox precipitates. Biosens Bioelectron 85:220–225
Acknowledgments
The authors are thankful for the support of this work by the National Natural Science Foundation of China (Grant No. 21575165), the Hunan Provincial Science and Technology Plan Project, China (No. 2019TP1001), and the Innovation-Driven Project of Central South University (2020CX002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 866 kb)
Rights and permissions
About this article
Cite this article
Lv, Z., Wang, Q. & Yang, M. DNAzyme-Au nanoprobe coupled with graphene-oxide–loaded hybridization chain reaction signal amplification for fluorometric determination of alkaline phosphatase. Microchim Acta 188, 7 (2021). https://doi.org/10.1007/s00604-020-04681-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-020-04681-1