Abstract
Various methods have been developed in recent years for the determination of uranyl ion by making use of uranyl-specific DNAzymes. However, many of them suffer from hydrolysis by nucleases present in samples such as body fluids. We report here on an uranyl-specific nuclease-resistant DNA aptamer (UApt) as the recognition element, and how gold nanoparticles (AuNPs) can be used as signal reporters in the respective assay. The presence of uranyl ion leads to a conformational change of UApt, and this results in the dispersion of AuNPs and a decrease in the intensity of resonance light scattering (RLS) at around 573.0 nm. The conformational changes were also studied by polyacrylamide gel electrophoresis, circular dichroism, and UV–vis spectroscopy. The RLS signals are linearly related to the concentration of uranyl ion in the 22 to 550 nM range, with a detection limit of 6.7 nM. This method is more simple and robust than others owing to use of a UApt without a ribonucleotide adenosine. It has been successfully applied to the determination of uranyl ion in real samples. We presume that this method may be extended to the determination of other analytes by making use of the corresponding aptamer for the target.

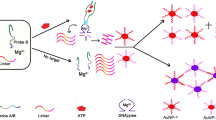
The presence of UO2 2+ leads to the conformation change of UApt, resulting in the dispersion of AuNPs and decrease of the RLS intensity of the system.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Uranium is an ubiquitous trace radioactive element that has received much attention owing to its high toxicity and radioactivity [1, 2]. Recently, low concentration of uranium has been widely detected in most of ground water, tap water, and drinking water [3]. The U.S. Environmental Protection Agency defined the maximum contamination level of UO2 2+ in drinking water to be 130 nM. Due to its growing usage in both civilian and military applications, human beings have a high chance of being exposed to uranium, which could threaten the human health [4, 5]. Therefore, highly selective and sensitive determination of trace uranium is of great significance in environmental protection and public security.
To date, many methods for the determination of uranium have been developed, such as inductively coupled plasma-mass spectrometry [6], X-ray fluorescence [7], time-resolved laser fluorescence spectroscopy [8], and capillary zone electrophoresis [9, 10]. However, it is generally considered those most of them either require sophisticated equipment and skilled operators, or involve tedious and painstaking procedures.
In recent years, DNAzymes as a new class of reporters have drawn great research interest worldwide owing to their biological importance and applications [11–13], which can be obtained through in vitro selection process from a large DNA library. So far, a number of DNAzymes have been selected, and used to prepare metal sensors based on their specific recognition to metal ions [11, 12, 14–16]. For example, Lu and co-worker reported a catalytic beacon for the detection of uranyl using an uranyl ion-specific DNAzyme [12], which consists of a DNA enzyme strand and a DNA substrate with a ribonucleotide adenosine (rA) in the middle. This strategy was demonstrated to have a 45 pM detection limit. Furthermore, they reported colorimetric methods for uranium based on uranyl (UO2 2+) specific DNAzyme and gold nanoparticles (AuNPs), which have advantages of high selectivity and simple instrument [11]. In our last work [16], we developed an assay of uranyl using a labeled DNAzyme-AuNPs system by a resonance light scattering (RLS) technique, which has some distinct advantages of simplicity, rapidness, and high sensitivity [17, 18]. Recently, DNAzyme based electrochemical sensors for trace uranium was also reported [19]. Although there has been remarkable progress in the development of these detection methods, the vulnerability of DNAzymes to nucleases and complicated operation for labeling DNAzyme are the drawbacks of DNAzymes-based strategies for the biomonitoring of target molecules in body fluids. It is well known that the DNAzymes are the DNA/RNA chimera with a ribonucleotide adenosine (rA), which was introduced to serve as the putative cleavage site for in vitro selection, because a ribonucleotide is about 100,000 fold more susceptible to hydrolytic cleavage than a deoxyribonucleotide [12, 20]. Thus, they are easily degradable by ribonuclease. Owing to the vulnerability of DNAzymes to nucleases, all the solutions used in experiment must be treated with 0.1 % diethyl pyrocarbonate (DEPC) or other nuclease suppressors to create and maintain the RNase-free environment [16, 21]. In addition, in most cases, an appropriate probe such as AuNPs or fluorophore is tethered to DNAzymes to indicate color or light intensity change [11, 12, 16]. Above those methods are tedious, time-consuming, and increase experimental cost. A number of research methods have attempted to overcome the instability of functional nucleic acids by using chemical modifications [22]. However, these methods cannot guarantee the preservation of the performance of the functional nucleic acids for detecting metals. Therefore, it is a substantial challenge to select specific DNA sequences for metal ions, and subsequently engineer them to allow the stable and sensitive detection of analytes. Fortunately, an uranyl specific nuclease-resistant DNA aptamer (UApt) has been recently reported, which can specifically bind to UO2 2+ with high affinity and stability [23]. This success prompts us to purposefully prepare a specific DNA aptamer by reasonable modification for UO2 2+ detection. To our knowledge, there is no report on the determination of uranium using UApt by RLS strategy.
In the present work, taking advantage of the high stability and specificity of UApt, we designed a label-free RLS method for the sensitive, selective determination of UO2 2+. For this purpose, an UApt is used for the recognition of UO2 2+, and AuNPs as signal reporter, which have high extinction, strong size- and distance-dependent properties [11, 24, 25]. The presence of UO2 2+ leads to the change of UApt conformation, resulting in the dispersion of AuNPs and the decrease of the RLS intensity of the system. Thereby, a novel RLS method for the determination of UO2 2+ was developed. As a proof-of principle experiment, we demonstrated that the developed RLS method can sensitively and selectively detect UO2 2+ in real samples. The detection limit obtained for UO2 2+ was 6.7 nM, which is below the maximum contamination level of 130 nM for uranium in drinking water defined by the U.S. Environmental Protection Agency. Furthermore, this detection system could be expanded easily to other targets by simply replacing corresponding aptamer. Thus, this new methodology can be expected to provide a highly sensitive platform for the analysis of various target molecules.
Experimental
Materials and chemicals
HPLC-purified oligonucleotides were purchased from Sangon Biotechnology Co., Ltd. (Shanghai, China, http://sangon.bioon.com.cn/). Their sequences were designed according to the literature with an appropriate modification [23] and shown as follow: DNA1, 5′-CAC GTC CAT CTC TGC AGT CGG GTA GTT AAA CCG ACC TTC AGA CAT AGT GAG T-3′; DNA2, 5′-ACT CAC TAT AGG AAG AGA TGG ACG TG-3′. HAuCl4 · 4H2O, sodium citrate, and uranium nitrate hexahydrate were purchased from Shanghai First Reagent Plant (Shanghai, China, http://2064257.atobo.com.cn/). Uranium nitrate hexahydrate was dissolved in water to make a 1 mM stock solution. All reagents were of analytical grade, and ultra-pure water (18.2 MΩ) was used throughout.
Apparatus
All RLS spectra were obtained with a Shimadzu F-5301 spectrofluorometer (Kyoto, Japan). A Shimadzu UV-2450 spectrophotometer (Kyoto, Japan) was used to measure the absorption spectra. The exact concentrations of DNA were checked by SmartSpec™ plus spectrophotometer. A MVS-1 vortex mixer (Beijing, China) was used to blend mixtures.
Preparation of gold nanoparticles
AuNPs were prepared according to the method described in our previous study [16]. Briefly, HAuCl4 of 1 mM was reduced to AuNPs by 5 mL of 38.8 mM sodium citrate. The resulting AuNPs were filtered through a 0.22 μm membrane filter and stored at 4 °C away from light. The diameter of above-prepared AuNPs was 13 ± 1.7 nm, and the concentration was about 8.1 nM according to Beer’s law using an extinction coefficient of 2.7 × 108 M−1 cm−1 at λmax 520 nm.
DNA hybridization
The oligonucleotides were dissolved in water and quantitated by SmartSpecTM plus spectrophotometer. The equal amounts of DNA1 and DNA2 were incubated for 3 min in 100 mM NaCl solution at 90 °C, then cooled to room temperature for 1 h to form DNA1-DNA2 duplex as an UO2 2+-specific DNA aptamer.
Procedure for resonance light scattering determination
RLS measurements were performed on a Shimadzu F-5301 spectrofluorometer. A 19 μL UApt was mixed with various concentrations of UO2 2+ or sample solutions in 100 μL of MES buffer solution (pH 5.5), and reacted for 6 min at room temperature. After addition of 50 μL of AuNPs, the mixture solution was diluted to 230 μL by water. Then, 170 μL of 200 mM NaCl was slowly added and shaken sufficiently. After that, the RLS intensity of the system was measured at λem = 573.0 nm, and the excitation and emission slit width were at 3.0 nm and 5.0 nm, respectively. The decreased RLS intensity of the system was represented as ΔI = I 0 − I, where I and I 0 were the RLS intensities of the system with and without UO2 2+, respectively.
Circular dichroism spectra determination
Circular dichroism (CD) spectra of the UO2 2+-specific DNA aptamer in the presence and absence of UO2 2+ in the MES buffer were measured on a JASCO J-815 circular dichroism spectrometer at room temperature. It was recorded from 200 nm to 370 nm in 1 mm pathlength cuvettes and averaged from five scans. The scanning speed was 500 nm per minute.
Polyacrylamide gel electrophoresis and sliver staining
Polyacrylamide gel electrophoresis assays were performed according to the reference with an appropriate adjustment [26]. The DNA solution mixed with 6× loading buffer was analyzed in 16 % polyacrylamide gel (acrylamide/bisacrylamide, 29:1) at constant voltage of 90 V for 130 min. 1×TBE (pH 8.3) was used as electrophoresis buffer. After that, the gel was dealt with argentation as follow: firstly, the gel was fixed via 10 % ethanol for 20 min and rinsed three times by deionized water. Then, it was stained by 0.2 % AgNO3 away from light and rinsed three times by deionized water. Finally, it was developed by the mixture solution of 3 % NaOH and 0.3 % formaldehyde.
Result and discussion
Design strategy and analytical principle
The analytical principle for this strategy can be stated that the binding of an UApt with target molecule forces the UApt to undergo a conformational change, which results in the change of the aggregation and dispersion states of AuNPs along with a change of RLS intensity of the system. Therefore, the strategy for designing RLS method can be summarized as how to transform the conformation change of an UApt to RLS intensity change of a system in the presence of UO2 2+. In order to achieve the mentioned above sensing performance, we designed a novel UApt for the determination of UO2 2+. As shown in Fig. S1 (see Supplementary data), the UApt was prepared by simply replacing rA in the reported UO2 2+-specfic DNAzyme with a deoxyribonucleotide adenosine or by removing bases at the 3′ and 5′ ends in the reported DNA aptamer for UO2 2+ [11–13, 23]. The AuNPs was employed as an RLS probe to indicate a change of RLS intensity of the system. The UApt (dsDNA) constructed through the hybridization between DNA1 and DNA2 was stiff and exposed a negatively charged phosphate backbone. The strong repulsion between UApt and AuNPs makes their binding negligible, causing salt-induced aggregation [11]. Upon addition of UO2 2+, the conformational change of the UApt results in the change of the states of AuNPs from aggregation to dispersion states along with a change of RLS intensity of the system (Scheme 1). Under the optimum experimental conditions, ΔI is directly proportional to the concentration of UO2 2+. Thereby, a novel method was established for the determination of UO2 2+ based on a linear relationship between the concentration of UO2 2+ and ΔI of system.
Mechanism of the interaction of aptamer with uranyl
As shown in Fig. 1, the I RLS of AuNPs-UApt solution was very high (curve 1). Upon addition of UO2 2+, the RLS intensity of the system at 339 nm and 573 nm gradually decreased (curves 2–6). Our previous experimental result demonstrated that the cleavage of substrate strand by DNAzyme in the presence of UO2 2+ resulted in releasing a single stranded DNA (ssDNA) [16]. To confirm whether the UApt has catalytic activity or not, we carried out a polyacrylamide gel electrophoresis assay. Interestingly, the experimental results were opposite to those obtained by an UO2 2+-specific DNAzyme. As shown in Fig. 1 (inset), no cleavage was observed after addition of UO2 2+ (lane 4), indicating that the UApt had no catalytic activity in the presence of UO2 2+. This result also prompts us that the UApt might undergo a conformational change in the presence of UO2 2+, which results in the change of RLS intensity of the system.
UV–vis spectra were used to testify the presumption mentioned above. It can be seen from Fig. 2a that the maximum absorption peak (λ max) of AuNPs oneself was located at 521 nm (curve 1). Upon addition of NaCl, the λ max of the system shifted to 534 nm, and the absorbance of the system also decreased notably (curve 2), indicating the formation of aggregates of AuNPs due to salt-induced screening effect [11]. When the UApt was added into the above-mentioned solution, the λ max changed from 534 nm to 524 nm, and the absorbance of the system was increased (curve 3), implying that UApt probably could interact with AuNPs to result in the dispersion of AuNPs aggregates partly. The most likely explanation of this result is that there may be a dynamic balance of a hairpin structure and a random coil sequence (unfolded single-stranded DNA) in the UApt -AuNPs-NaCl solution, while UApt mainly existed in a hairpin structure (dsDNA). It is well known that the hairpin structure (dsDNA) is much rigider than the ssDNA and ultimately prevent the exposure of the DNA bases to the AuNPs. Therefore, it cannot be adsorbed on AuNPs and lose the ability of protecting the AuNPs, causing salt-induced aggregation. On the other hand, a little ssDNA could be adsorbed onto AuNPs to result in the disassembly of the AuNPs aggregates, leading to the increase of the absorbance at 524 nm. Upon addition of UO2 2+, the value of absorbance was further increased at 524 nm. According to previous reports [11], an increase of the extinction indicates the melting of the hybridized DNA. Herein, we presumed that the interaction of the UApt with UO2 2+ may result in the disassembly of the AuNPs aggregates due to the dehybridization of the UApt which was constructed by DNA1 and DNA2, leading to the increase of the absorbance at 524 nm.
CD spectra were also used to confirm above mentioned presumption. As shown in Fig. 2b, the CD spectra of the UApt in MES buffer alone displayed two positive bands at 214.3 nm and 272.3 nm due to base stacking, and a negative band at 239.5 nm due to polynucleotide helicity. This result indicates that UApt mainly existed in a hairpin structure (dsDNA) [27, 28], which is a bulged stem-loop structure with a number of mismatches (see Supplementary data, Fig. S1). Upon addition of UO2 2+, the positive band at 214.3 nm shifted to 217.3 nm along with a decrease of the intensity, indicating the existence of single stranded DNA (ssDNA) [29]. Whereas the negative band at 239.5 nm and the positive band at 272.3 showed minor red shift of 1 nm along with an increase in intensity. These results suggested that the interaction of UApt with UO2 2+ results in the conformational change of UApt, which was very important to the RLS intensity change of the system. Based upon above-mentioned phenomena and literatures [11, 12, 20, 27], the mechanism of the assay could be explained as following: UApt mainly existed in a hairpin structure (dsDNA). Upon addition of UO2 2+, the interaction of an UApt with UO2 2+ forces the UApt to undergo a conformational change, which results in the change of the aggregation and dispersion states of AuNPs along with a change of RLS intensity of the system. According to previous report [20], the uranyl (UO2 2+) ion has a high affinity towards the phosphate oxygen of UApt as it is a hard Lewis acid. So, it can react with UApt to form a complex, and result in the disruption of DNA base-pairing interactions by UO2 2+/UApt interactions, that is, the interaction between the uranyl ion and UApt might result in the dehybridization of the UApt to form ssDNA, which could be adsorbed onto AuNPs to prevent them from aggregating in the presence of NaCl, leading to the decrease of the RLS intensity of the system. Therefore, the I RLS of system decreased gradually with increasing UO2 2+ concentration.
Optimization of experimental conditions
In this strategy, the effect of pH on the RLS intensity was conducted in the pH range of 4.5–6.5 using 10 mM MES. As shown in Fig. 3a, the RLS intensity was enhanced rapidly in the pH range of 4.5–5.25, and remained relatively steady in the pH range of 5.25–5.75, with a maximum value of pH 5.5. Besides, the RLS intensity of the system decreased rapidly in the pH range of 5.75–6.50. The most likely explanation of such experimental result might be that pH influences the uranyl ion speciation in solution, and an uranyl species was produced at pH 5.5, which is the most effective species for carrying out the reaction. When pH values are higher than 5.5, hydrolyzed species such as UO2(OH)+ start to dominate [20]. At pH > 7, a sediment of uranyl can be observed, which was also unfavorable for the interaction of UO2 2+ with UApt. Therefore, MES buffer solution of pH 5.5 was selected for controlling the acidity of the solution.
The influence of the adding volumes of AuNPs on the RLS intensity of the system was also studied in the range of 40 ~ 60 μL. The results showed that the RLS intensity was remarkably enhanced in the range of 40 ~ 50 μL, with a maximum value of 50 μL, and decreased in the range of 50 ~ 60 μL (Fig. 3b). The reason may be that the interaction between the uranyl ion and UApt might result in the dehybridization of the UApt to form ssDNA. The resulting ssDNA can easily be adsorbed on AuNPs and enhance electrostatic repulsion between AuNPs, which in turn stabilizes AuNPs even in the presence of high salt concentration. With the increase of the concentration of AuNPs, the number of ssDNA on each AuNPs is reduced accordingly [30], that is, the ssDNA densities on each AuNPs were decreased. This causes salt-induced aggregation, resulting in the increase of the RLS intensity of the system with uranyl ions. Therefore, the ΔI value of the system was decreased. So, 50 μL of AuNPs was chosen for the further experiment.
The effect of temperature on RLS intensity of the system was examined every 10 °C in the range of 15–55 °C. The experimental results indicated that the RLS intensity of the system remained relatively stable in the range of 15–25 °C, and decreased gradually with increasing temperature in the range of 25–55 °C (Fig. 3c). The possible reason may be that a higher incubation temperature resulted in the heat denaturation of the DNA duplex to form ssDNA, which can be adsorbed on AuNPs via the stronger coordination interaction between the N atoms of the ssDNA and the AuNPs to protect the AuNPs from salt-induced aggregation, leading to decreasing the blank value of the system. Therefore, the ΔI values of the system decreased accordingly. So, the experiments were carried out at room temperature.
Selectivity of the resonance light scattering method
The effect of the potentially interfering ions on the determination of uranyl was investigated by analyzing synthetic sample solutions containing 50 nM UO2 2+, where a series of metal ions such as Zn2+, Cd2+, Pb2+, Ca2+, Fe2+, Fe3+, Mn2+, Hg2+, K+ and inorganic anions including PO4 3−, H2PO4 −, HPO4 2− had been added. The allowable quantities of the potentially interfering ions were defined as an error not more than ±3 %. The experiment results demonstrated that 800 times of K+, 500 times of PO4 3−, 400 times of Zn2+ and H2PO4 −, 200 times of Hg2+, 100 times of Cd2+ and Mn2+, 80 times of Ca2+, 50 times of Fe2+ and Pb2+, 30 times of Fe3+, 20 times of HPO4 2− do not interfere with determination. It was observed that a lot of metal cations and inorganic anions may be tolerated at high concentration. So the selectivity of the proposed method was better than other methods such as catalytic beacon [12].
Analytical parameters
Under the optimal condition, the sensitivity of the method was investigated. Fig. S2 (see Supplementary data) shows a good linear correlation (r = 0.9889) between the ΔI value and the concentration of UO2 2+ ion over the range of 22 to 550 nM, and the limit of detection was 6.7 nM for UO2 2+ calculated by the equation LOD = 3S b/m, where S b is the standard deviation of the blank measurements (n = 11) and m is the slope of the calibration graph. The detection limit of this method is below the maximum contamination level of 1.30 × 10−7 M for uranium in drinking water defined by the U.S. Environmental Protection Agency. The equation of linear regression was ΔI RLS = 70.9 + 4.10 c (×10−8 M).
Analysis of uranyl in real samples
To investigate whether this method was applicable to natural samples, seven samples were collected from the locations near an uranium mine, including the uranium ore leaching solution, the surface water of an uranium mine, and the pond water near an uranium mine, and the Xiangjiang river near the outlet of an uranium mine factory. The high concentrations of samples 1 and 2 were diluted for 1,000 and 500 times by ultra-pure water, respectively. The low concentrations of samples 3 to 7 were preconcentrated for ten times. All samples were filtrated, and then tested by the developed strategy. Recovery test was carried out by adding a known amount of UO2 2+ ions to samples, and the results were shown in Table 1. The results gained from both this strategy and the DNAzyme-based method [16] were statistically analyzed using the Student t-test, which do not exceed the theoretical value at 95 % confidence level, indicating no statistically difference between the two methods. It is obvious that the developed method for UO2 2+ detection was reliable, practical, and economical in its operation.
Comparison of this strategy with other methods
In this work, we achieve an unlabeled strategy for RLS detection of UO2 2+ ions using UApt as recognition element. Compared with the method based on labeled DNAzyme system [12, 16, 21, 19] and free-labeled nanogold catalytic method [14], this strategy has several excellent features. Firstly, all the solutions used in experiment do not need treating by DEPC owing to use of nuclease-resistant UApt. In addition, the assay does not involve any chemical modification of DNA. Thus, the assay is obviously more convenient and economical than the other methods with DNAzymes or aptamer labeled by AuNPs or fluorophore [16, 25]. Second, the synthesis of UApt without a ribonucleotide adenosine (rA) is easier than that of DNAzymes, which also decreases largely the experimental cost. Compared with inductively coupled plasma-mass spectrometry (ICP-MS) or atomic emission spectrometry (AES) [6, 31], our method without tedious procedure or the requirement of sophisticated equipment, is also more convenient and economical. Furthermore, as shown in Table S1 (see Supplementary data), this assay has a lower detection limit of 6.7 nM, which is better than other methods such as capillary zone electrophoresis [9], time-resolved laser fluorescence spectroscopy [8], chelating polymeric sorbent-spectrophotometry [32], and UV–vis [33]. Finally, as a substitute, UApt is the other functional DNA molecule for sensing UO2 2+, which was demonstrated to be able to overcome the drawback of instability of the DNAzymes owing to the nuclease-resistant characteristic of UApt. Therefore, this strategy can be expected to expand to other DNAzymes by simply replacing ribonucleotides in the DNAzymes with deoxyribonucleotides, and provide a highly sensitive platform for the analysis of various target molecules.
Conclusions
In summary, we have developed a rapid, label-free, selective and sensitive method for the detection of UO2 2+, using the UO2 2+ specific nuclease-resistant DNA aptamer as recognition element for the first time. This reliable sensing platform abrogates the tedious and painstaking procedures, and does not need any chemical modification of DNA probe and sophisticated equipment, thus decreasing largely the experimental cost and simplifying experimental procedure. This work has been successfully applied for detecting UO2 2+ in real samples with a satisfactory result. In addition, the mechanism of the interaction of UApt with UO2 2+ was investigated by polyacrylamide gel electrophoresis, CD, and UV–vis spectra, which is benefit to extend the application of UApt.
References
Darolles C, Broggio D, Feugier A, Frelon S, Dublineau I, De Meo M, Petitot F (2010) Different genotoxic profiles between depleted and enriched uranium. Toxicol Lett 192:337–348
Pereira S, Camilleri V, Floriani M, Cavalié I, Garnier-Laplace J, Adam- Guillermin C (2012) Genotoxicity of uranium contamination in embryonic zebrafish cells. Aquat Toxicol 109:11–16
Nriagu J, Nam DH, Ayanwola TA, Dinh H, Erdenechimeg E, Ochir C, Bolormaa TA (2012) High levels of uranium in groundwater of Ulaanbaatar, Mongolia. Sci Total Environ 414:722–726
Banday AA, Priyamvada S, Farooq N, Yusufi AN, Khan F (2008) Effect of uranyl nitrate on enzymes of carbohydrate metabolism and brush border membrane in different kidney tissues. Food Chem Toxicol 46:2080–2088
Shaki F, Hosseini MJ, Ghazi-Khansari M, Pourahmad J (2012) Toxicity of depleted uranium on isolated rat kidney mitochondria. Biochim Biophys Acta Gen Subj 1820:1940–1950
Boulyga SF, Becker JS, Matusevitch JL, Dietze HJ (2000) Isotope ratio measurements of spent reactor uranium in environmental samples by using inductively coupled plasma mass spectrometry. Int J Mass Spectrom 203:143–154
Misra NL, Dhara S, Óvári M, Záray G, Aggarwal SK, Varga I (2010) Determination of low atomic number elements at trace levels in uranium matrix using vacuum chamber total reflection X-ray fluorescence. Spectrochim Acta B 65:457–460
Lehmann S, Geipel G, Grambole G, Bernhard G (2009) A novel time-resolved laser fluorescence spectroscopy system for research on complexation of uranium(IV). Spectrochim Acta A 73:902–908
Sladkov V, Fourest B (2009) Simultaneous determination of uranium carbide dissolution products by capillary zone electrophoresis. J Chromatogr A 1216:2605–2608
Sladkov V, Zhao Y, Mercier-Bion F (2011) Capillary zone electrophoresis for U(VI) and short chain carboxylic acid sorption studies on silica and rutile. Talanta 83:1595–1600
Lee JH, Wang Z, Liu J, Lu Y (2008) Highly sensitive and selective colorimetric sensors for uranyl (UO22+): Development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J Am Chem Soc 130:14217–14226
Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y (2007) A catalytic beacon sensor for uranium with parts-pertrillion sensitivity and millionfold selectivity. Proc Natl Acad Sci U S A 104:2056–2061
Liu J, Cao Z, Lu Y (2009) Functional nucleic acid sensors. Chem Rev 109:1948–1998
Jiang Z, Zhang Y, Liang A, Chen C, Tian J, Li T (2012) Free-labeled nanogold catalytic detection of trace UO22+ based on the aptamer reaction and gold particle resonance scattering effect. Plasmonics 7:185–190
Xiang Y, Lu Y (2011) Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat Chem 3:697–703
Zhou B, Shi LF, Wang YS, Yang HX, Xue JH, Liu L, Wang YS, Yin JC, Wang JC (2013) Resonance light scattering determination of uranyl based on labeled DNAzyme–gold nanoparticle system. Spectrochim Acta A 110:419–424
Fan Y, Long YF, Li YF (2009) A sensitive resonance light scattering spectrometry of trace Hg2+ with sulfur ion modified gold nanoparticles. Anal Chim Acta 653:207–211
Zou QC, Zhang JZ, Chai SG (2011) Resonance light scattering method for the determination of DNA with cationic methacrylate based polymer nanoparticle probes. Spectrochim Acta A 82:437–443
Tang Q, Yuan Y, Xiao X, Hu J, Ma D, Gao Y (2013) DNAzyme based electrochemical sensors for trace uranium. Microchim Acta 180(11–12):1059–1064
Brown AK, Liu J, He Y, Lu Y (2009) Biochemical characterization of a uranyl ion-specific DNAzyme. ChemBioChem 10:486–492
Yin BC, Zuo P, Huo H, Zhong X, Ye BC (2010) DNAzyme self-assembled gold nanoparticles for determination of metal ions using fluorescence anisotropy assay. Anal Biochem 401:47–52
Wang L, Yang CYJ, Medley CD, Benner SA, Tan WH (2005) Locked nucleic acid molecular beacons. J Am Chem Soc 127:15664–15665
Kim J, Kim MY, Kim HS, Hah SS (2011) Binding of uranyl ion by a DNA aptamer attached to a solid support. Bioorg Med Chem Lett 21:4020–4022
Miao XM, Ling LS, Shuai XT (2012) Detection of Pb2+ at attomole levels by using dynamic light scattering and unmodified gold nanoparticles. Anal Biochem 421:582–586
Xu Z, Huang X, Dong C, Ren J (2013) Fluorescence correlation spectroscopy of gold nanoparticles, and its application to an aptamer-based homogeneous thrombin assay. Microchim Acta. doi:10.1007/s00604-013-1132-2
Zhu J, Li T, Zhang L, Dong S, Wang E (2011) G-quadruplex DNAzyme based molecular catalytic beacon for label-free colorimetric logic gates. Biomaterials 32:7318–7324
Gou XC, Liu J, Zhang HL (2010) Monitoring human telomere DNA hybridization and G-quadruplex formation using gold nanorods. Anal Chim Acta 668:208–214
Rajesh J, Gubendran A, Rajagopal G, Athappan P (2012) Synthesis, spectra and DNA interactions of certain mononuclear transition metal(II) complexes of macrocyclic tetraaza diacetyl curcumin ligand. J Mol Struct 1010:169–178
Zheng B, Cheng S, Liu W, Lam MHW, Liang H (2012) A simple colorimetric pH alarm constructed from DNA–gold nanoparticles. Anal Chim Acta 741:106–113
Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y (2008) DNA aptamer folding on gold nanoparticles: from colloid chemistry to biosensors. J Am Chem Soc 130:3610–3618
Purohit PJ, Goyal N, Thulasidas SK, Page AG, Sastry MD (2000) Electrothermal vaporization – inductively coupled plasma-atomic emission spectrometry for trace metal determination in uranium and thorium compounds without prior matrix separation. Spectrochim Acta B 55:1257–1270
Jain VK, Pandya RA, Pillai SG, Shrivastav PS (2006) Simultaneous preconcentration of uranium(VI) and thorium(IV) from aqueous solutions using a chelating calix[4]arene anchored chloromethylated polystyrene solid phase. Talanta 70:257–266
Ozdemir S, Kilinc E (2012) Geobacillus thermoleovorans immobilized on Amberlite XAD-4 resin as a biosorbent for solid phase extraction of uranium (VI) prior to its spectrophotometric determination. Microchim Acta 178(3–4):389–397
Acknowledgments
The authors gratefully acknowledge the support of the National Natural Science Foundation of China (No. 21177052), the Science and Technology Program of Hunan Province in China (No. 2010SK3039) and the Construct Program of the Key Discipline (Public Health and Preventive Medicine) in Hunan Province.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 149 kb)
Rights and permissions
About this article
Cite this article
Zhou, B., Wang, YS., Yang, HX. et al. A sensitive resonance light scattering assay for uranyl ion based on the conformational change of a nuclease-resistant aptamer and gold nanoparticles acting as signal reporters. Microchim Acta 181, 1353–1360 (2014). https://doi.org/10.1007/s00604-014-1267-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-014-1267-9