Abstract
We describe a sensitive chronocoulometric biosensor for the sequence-specific detection of DNA. It is based on a glassy carbon electrode modified with multi-walled carbon nanotubes, polydopamine, and gold nanoparticles. The ruthenium(III)hexammine complex acts as the electrochemical indicator. Electrochemical impedance spectra and scanning electron microscopy are employed to investigate the assembly of the electrode surface. The signals of the ruthenium complex electrostatically bound to the anionic phospho groups of the DNA strands are measured by chronocoulometry before and after hybridization. The difference in signal intensity is linearly related to the logarithm of the concentration of the target DNA in the range of 1.0 nM to 10 fM with a detection limit of 3.5fM (S/N = 3) under optimal conditions. This biosensor exhibits excellent sensitivity and selectivity and has been used for an assay of complementary target DNA in human serum sample with satisfactory results.

We describe a sensitive chronocoulometric biosensor based on a glassy carbon electrode modified with gold nanoparticles, poly(dopamine), and carbon nanotubes. The biosensor exhibits excellent sensitivity and selectivity and has been used for an assay of Helicobacter pylori in human serum with a satisfactory result.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Sequence-specific detection of DNA targets associated with genetic or pathogenic diseases has become increasingly important in molecular diagnostics. Various techniques have been developed for such detection, including optical [1, 2], surface-enhanced Raman scattering [3], electrochemistry [4–10] and piezoelectric transducers [11, 12]. Among these techniques electrochemical biosensors have attracted considerable attention due to their simplicity, low cost and high sensitivity. Up to date, many electrochemical approaches to detecting DNA have been reported. For example, Fang and his co-workers reported a series of highly sensitive electrochemical DNA biosensors based on nanoparticles and carbon nanotubes composite modified electrodes [13, 14]; Yu and his co-workers have reported molecular beacon-based electrochemical DNA biosensors [15, 16]. Fan and Zhang’s groups have fabricated several chronocoulometric DNA sensors with hexaammineruthenium (III) chloride acting as indicator [17–19], which can detect fM target DNA; Jiao and his co-workers have constructed electrochemical DNA biosensors based on a poly (2,6-pyridinedicarboxylic acid) film modified electrode [20, 21]; Later they fabricated a sensitive DNA biosensor composed of polyaniline nanofibers and MWCNTs composites film, which can detect 27 fM target DNA [22]. Recently, nanoparticles and polymers are often used in sensing fields to improve sensor performance owing to their advantages (e.g. higher surface area and conductivity), and some reviews have been published about polymers or nanoparticles in sensing field application [23–25].
Dopamine (2-(3,4-dihydroxyphenyl)ethylamine) (DA) is one of the neurotransmitters, whose level in the body is related to some diseases. It is easily oxidized and polymerized at the electrode surface. A previous study shows that the DA polymer film has good electrochemical response for some biomolecules [26] and that the film has been used in electrochemical biosensors. For example, Wang and co-workers employed poly(dopamine) film to fabricate an electrochemical immunosensor for the detection of interleukin-6 amperometric [27].
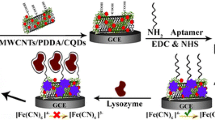
As is well known, multi-walled carbon nanotubes (MWCNTs) have unique properties (e.g. high surface area and good conductivity), and gold nanoparticles (Au-NPs) have good biocompatibility. Therefore, MWCNTs and Au-NPs have been extensively used as sensing materials for the preparation of sensors. Inspired by the DA polymer film advantages and MWCNTs properties, we describe the fabrication of a sensitive DNA biosensor. In this study, MWCNTs, DA polymer film and Au-NPs were used as sensing interface. Chronocoulometry was utilized to investigate the DNA hybridization process, and hexaammineruthenium (III) chloride (RuHex) was used as the indicator. Previous studies have demonstrated that the bindings of RuHex to DNA are completely through electrostatic interaction, while free of any duplex intercalation, and the bindings to anionic phosphate of DNA strands are a stoichiometric approach [28, 29]. Hence, the redox charge of RuHex is a direct function of the amounts of DNA strands localized on electrode surfaces. In our study, MWCNTs and DA polymer film clearly enhanced the electrochemical response of the RuHex. As a result, the biosensor shows higher sensitivity and better selectivity, which can distinguish single-base mismatched target DNA. The fabrication of the biosensor and detection of DNA is shown in Scheme 1.
Experimental
Chemicals and reagents
Chloroauric acid (HAuCl4·4H2O) and HCl were obtained from Shanghai Chemical Reagent Co. Ltd (Shanghai, China, http://ccn.mofcom.gov.cn). Dopamine (DA), sodium dodecyl sulfate (SDS), Tris-(hydroxymethyl) aminomethane (Tris-base) and hexaammineruthenium (III) chloride ([Ru(NH3)6]3+, RuHex) were purchased from Alfa Aesar Co. Ltd (Tianjin, China, http://www.alfa.com). Multi-walled carbon nanotubes with carboxylic acid groups (MWCNTs, with a diameter of about 20–30 nm and an approximate length of 30 μm) were obtained from Chengdu Institute of Organic Chemistry, Chinese Academy of Sciences, and used without further purification (http://www.timesnano.com). All oligonucleotide fragments used were purchased from Shanghai Sangon Inc. (Shanghai, China, http://www.sangon.com). Their base sequences were as follows:
-
Probe sequences: 5′-SH-(CH2)6-GCG-TTC-CAA-AGG-GCA-GGA-TCA-TTG-A-3′
-
Complementary sequences: 5′-TCA-ATG-ATC-CTG-CCC-TTT-GGA-ACG-C-3′
-
Non-complementary sequences: 5′-AGC-CCA-CAC-TGA-TGG-CGC-CAC-TGC-A-3′
-
Single-base mismatched sequences: 5′-TCA-ATG-ATC-CTG-TCC-TTT-GGA-ACG-C-3′
-
5′-TCA-ATG-ATC-CTG-GCC-TTT-GGA-ACG-C-3′
All oligonucleotide stock solutions were prepared with 10 mM Tris-HCl + 1 mM EDTA (pH 7.4) and were stored in a refrigerator. DA solution was prepared with 0.01 M phosphate buffered saline (PBS, pH 7.0). The buffers employed in this study were as follows: 0.01 M PBS (0.1 M NaCl + 0.01 M PBS, pH 7.0); 10 mM Tris-HCl (10 mM Tris-HCl + 0.25 M NaCl, pH 7.4). All solutions were prepared with twice-quartz-distilled water.
Apparatus
All electrochemical measurements were performed on a CHI 650C electrochemical workstation (Shanghai Chenhua Instruments Co., China) with a conventional three-electrode system composed of a platinum auxiliary, silver-silver chloride (Ag/AgCl) (with 3.0 M KCl) reference, and a bare GCE or modified GCE working electrode. All potentials in this study are referenced to the Ag/AgCl.
Electrochemical impedance spectra (EIS) were performed in 0.1 M PBS containing 5.0 mM [Fe(CN)6 3−/4−] at a pH of 7.4. The frequency ranged from 0.1 to 100 kHz at a formal potential of 0.18 V, and the amplitude of the alternate voltage was 5 mV.
All measurements were performed in a 10 mL electrolytic cell with 5.0 mL solutions, from which oxygen was removed by purging with high-purity nitrogen for 10 min, and a blanket of nitrogen was maintained over the solution during the measurements. All measurements were performed at room temperature. Scanning electron microscopy (SEM) (Hitachi JEOLJSM-6700 F, Japan) was used to obtain the morphology of different modified electrodes.
Preparation of the probe DNA modified electrode
Before modification the bare glassy carbon electrode (GCE) (3.0 mm in diameter, CH Instruments Inc.) was freshly polished with 1.0, 0.3 and 0.05 μm alumina powder, respectively. The electrode was then cleaned ultrasonically sequentially in 95 % ethanol and water for 3.0 min. Finally, the electrode was electrochemically cleaned between − 0.3 and + 1.5 V in 0.5 M sulfuric acid by repetitive cyclic potential scan for 20 cycles.
1.0 mg MWCNTs were dispersed in 10.0 mL of anhydrous ethanol with the aid of ultrasonic agitation to obtain a black suspension, and 5.0 μL of MWCNTs suspensions was dropped on the surface of the pretreated glassy carbon electrode and dried naturally at room temperature to form MWCNTs thin film. DA was then electropolymerized on the surface of the electrode modified with MWCNTs by repetitive cyclic potential scan for 10 cycles between − 0.7 and + 0.7 V at 100 mV·s−1 in 0.01 M PBS containing 2.0 mM DA. The modified electrode was denoted as PDA/MWCNTs/GCE.
Finally, the modified electrode mentioned above was immersed in 0.1 M NaNO3 solution containing 1.0 mg·mL−1 HAuCl4 and electrochemically deposited for 20 s at − 200 mV. Thus, Au-NPs were deposited on the surface of the electrode modified with PDA/MWCNTs and it was denoted as Au-NPs/PDA/MWCNTs/GCE.
Immobilization of the probe DNA was achieved by dropping 4.0 μL of the 1.0 × 10−5 M probe DNA onto the surface of the Au-NPs/PDA/MWCNTs-modified electrode, which was stored in a refrigerator for 1 h at 4 °C. Afterwards, the modified electrode was immersed in 0.1 % SDS for 10 min to remove mobile probe DNA. The obtained electrode was denoted as ssDNA/Au-NPs/PDA/MWCNTs/GCE.
Electrochemical detection of DNA hybridization
The DNA hybridization process was performed by immersing the probe-modified electrode in 0.01 M Tris-HCl buffer containing various concentrations of target DNA for 40 min at 37 °C. Then, the modified electrode was rinsed with 10 mM Tris-HCl 3 times to remove the unhybridized target DNA.
The DNA hybridization was assessed with the chronocoulometry technique by measuring the difference of the signal intensity of RuHex before and after hybridization with target DNA in 10 mM Tris-HCl (pH 7.4) containing 50 μM RuHex, and the concentration of complementary DNA was quantified by the difference in signal intensity of RuHex (ΔQ) (ΔQ = Qds−DNA − Qss−DNA). The experiment parameters are listed as follows: initial potential, 0.2 V; final potential, − 0.5 V; number of steps, 2; pulse width, 0.25 s; sample interval, 0.002; sensitivity (C or A/V): 5e–5 A/V.
Results and discussion
Electropolymerization of dopamine at the surface of the electrode modified with multi-walled carbon nanotubes
DA is an important neurotransmitter, whose electrochemical behavior has been supposed to occur through three-step reactions as shown in Scheme 2 according to previous literature [30]. In Scheme 2, the “E” refers to the electrochemical reactions and the “C” to the chemical reactions. The repetitive cyclic voltammograms were obtained in 2.0 mM DA at the MWCNTs/GCE modified electrode (See supplementary, Fig. S1). In the first positive scan, an oxidation peak (1) at ca. + 0.21 V was observed, which is attributed to the oxidation of the DA. In the subsequent reversal scan, two reduction peaks (2, 3) were observed at ca. + 0.1 V and − 0.3 V, which may be attributed to the reduction of dopaminequinone (DAQ) and dopaminechrome (DAC) as well as the intramolecular cyclization reaction product of dopaminequinone (DAQ). In the second positive scan, a new oxidation peak (4) appeared at ca. − 0.25 V, which may be attributed to the oxidation of leucodopaminechrome (LDAC). Simultaneously, the peak current of peak (1) and peak (2) decreased cycle by cycle, and the peak current of peak (3) and peak (4) continuously increased with continuous scanning, reflecting the continuous growth of the film. When the cyclic potential scan reached 10 cycles, the peak currents hardly changed. These facts indicate that DA was successfully polymerized on the surface of the electrode modified with MWCNTs by the electropolymerization mode. The polymerization mechanism of DA is described as follows: the part of dopaminechrome (DAC) molecules may evolve to the more stable 5,6-dihydroindole molecules of two aromatic rings via isomerization, and 5,6-indolequinone is then electropolymerized to form polymers (see Scheme 2).
Subsequently, the obtained PDA/MWCNTs modified electrode was immersed in 0.1 M NaNO3/1.0 mg·mL−1 HAuCl4 and electrodeposited for 20 s at − 200 mV. The obtained electrode was denoted as Au-NPs/PDA/MWCNTs/GCE.
Investigation of the electrode surface assembly process
SEM was used to observe the surface figures of the various modified electrodes. The SEM images of the MWCNTs (A), PDA/MWCNTs (B) and Au-NPs/PDA/MWCNTs film (C) are shown in Fig. 1. Figure 1a clearly shows that the MWCNTs are distributed very homogeneously on the surface of GCE. The image of Fig. 1b is obviously cloudy because the surface of the MWCNTs was covered with a thin film of DA. After Au-NPs had been electrodeposited on the surface of the PDA/MWCNTs film, it could be observed that the bright spots (Au-NPs) were homogeneously dispersed on the surface of the PDA/MWCNTs-modified electrode (Fig. 1c) (the diameter of the Au-NPs was about 50 nm). The changes in the SEM images confirmed the occurrence of the assembly process on the electrode surface.
EIS was used to investigate the changes of the electrode behavior after each assembly step. The impedance spectra included a semicircle portion at higher frequencies corresponding to the electro-transfer-limited process and a linear portion at lower frequencies representing the diffusion-limited process. The diameter of semicircle represents the electro-transfer resistance, Rct, which dominates the electron transfer kinetics of the redox probe at the electrode interface. Figure 2 exhibits the Nyquist plots of 5.0 mM [Fe(CN)6]3−/4− obtained on the electrodes surface after each assembly step. The inset is the electrical equivalent circuit. It could be observed that modification of the electrode surface strongly influences the diameter of the semicircle, thus leading to a change in the value of Rct, which makes it is possible to monitor each step of the sensing fabrication just following the variation of the parameter. For the bare GCE (curve a) and MWCNTs/GCE (curve b), the value of Rct was small (ca. 191 Ω). After DA was electrodeposited on the surface of MWCNTs/GCE, the value of Rct evidently increased from 191 Ω to 460 Ω (curve c) due to the polymer film block of the electron transfer of the [Fe(CN)6]3−/4−between the electrode surface and solution. After Au-NPs was introduced to the surface of the PDA/MWCNTs/GCE (curve d), the value of Rct obviously decreased from 460 Ω to 152 Ω because Au-NPs increases the conductivity of the electrode interface and improves the electron transfer rate of [Fe(CN)6]3−/4− at the electrode surface. Subsequently, the probe DNA was assembled on the surface of the Au-NPs/PDA/MWCNTs/GCE, the value of Rct rose from 152 Ω to 839.8 Ω due to the electrostatic repulsion between the negatively charged phosphate groups of DNA skeleton and the negatively charged redox probe ([Fe(CN)6]3−/4−)(curve e). After the target DNA hybridized with probe DNA to form the dsDNA, the value of Rct continuously increased to 1480.5 Ω (curve f) due to the improved quantity of negative charges. Hence, DNA hybridization can be proved by this further increment of electron transfer value.
Nyquist plots of 5.0 mM [Fe(CN)6 3−/4−] in 0.1 M PBS at different modified electrodes: bare GCE (a); MWCNTs/GCE (b); PDA/MWCNTs/GCE (c); Au-NPs/PDA/MWCNTs/GCE (d); ssDNA/Au-NPs/PDA/MWCNTs/GCE (e); dsDNA/Au-NPs/PDA/MWCNTs/GCE (f). Inset: Equivalent electrical circuit diagrams for the impedance plots. RCT: electron transfer resistance; Rs: electrolyte resistance; CDL: interface capacitance
In order to investigate the role of PDA and MWCNTs in sensing, we fabricated four different substrate biosensors (PDA, MWCNTs, PDA/MWCNTs, GCE) with the other conditions remaining constant (e.g. deposition and hybridization time), and selected signal intensity of RuHex as judge standard. The results are shown in Fig. S2 with histograms (the grayish bar represents the signal of the probe, and the black bar represents the signal of hybridization). It can be observed that the signal was weaker when the biosensor was fabricated with only MWCNTs or PDA, while it was strongest when the biosensor was fabricated with PDA/MWCNTs film, indicating that PDA/MWCNTs film enhanced the response signal.
Optimization of assay conditions
In this study, we investigate the effect of the assay conditions on the electrochemical signal (e.g. deposition time, amount of probe DNA and hybridization time). The results are shown in Fig. S3. Figure S3A displays the effect of Au-NPs on the signal of RuHex. It was found that the signal intensity of RuHex increased gradually with the increase of the deposition time from 5 to 25 s and reached the maximum value at 20 s. Hence, 20 s was selected for the deposition time. Figure S3B shows the influence of the probe DNA on the signal of RuHex. It was observed that the response signal of RuHex was strongest when 4.0 μL of probe DNA was employed. The reason is that excessive probe DNA will generate greater steric hindrance and reduce the hybridization efficiency. So we selected 4.0 μL probe DNA in this study. Similarly, the influence of the hybridization time on the electrochemical signal of RuHex was investigated (Fig. S3C). The signal of RuHex increased dramatically with the hybridization time from 0 to 30 min and remained nearly constant when the hybridization time was more than 40 min. Therefore, 40 min was employed as hybridization time in this study.
Selectivity of the DNA sensor
For DNA biosensors, selectivity is obviously a crucial factor to be considered. In this study, the selectivity of the DNA sensor was investigated using non-complementary, single-base mismatched and complementary sequences. Figure 3 displays histograms of the signal intensity of RuHex vs. different oligonucleotide sequences. It could clearly be seen that the signals obtained were different when this DNA biosensor recognized different oligonucleotide sequences. The response signal obtained from complementary sequences was the strongest and the signal obtained from non-complementary sequences was the lowest. The signal intensity of single-base mismatched sequences was about 25 % of that of complementary sequences, indicating that the DNA biosensor has good selectivity, enabling it to recognize single-base mismatched target DNA from complementary and non-complementary sequences.
a Chronocoulometry curves of RuHex after the probe (a) modified electrode hybridized with non-complementary sequences (b); single-base mismatched sequences of G or T (c, d); complementary sequences (e). b The histograms of the signal intensity of RuHex vs. different oligonucleotide sequences. Support electrolyte: 10 mM Tris-HCl +0.25 M NaCl (pH 7.4) containing 50 μM RuHex. The concentration of complementary target DNA is 1.0 × 10−9 M
Analytical performance
The analytical performance of this DNA biosensor was evaluated using a probe-modified electrode to hybridize with the different concentrations of the complementary target DNA under the optimal conditions. The results are shown in Fig. 4. The signal intensity of RuHex increased as the concentrations of the complementary target DNA increased, and it was linear to the logarithm of the concentration of the complementary target DNA from 10 fM to 1.0 nM. The linear regression equation was ΔQ = 5.388 lgCDNA + 81.69 (unit of C is M, unit of Q is μC), and the regression coefficient (R) of the linear curve was 0.9963. The detection limit was 3.5 fM (S/N = 3). Compared with similar work already published [31–35] (see Table 1), the biosensor has a wider linear range and lower limit of detection, but its sensitivity is lower than that of the “sandwich” type biosensor reported previously [19].
Chronocoulometry curves of the probe-modified electrode hybridized with target DNA in various concentrations (M): a 0; b 1.0 × 10−14; c 1.0 × 10−13; d 1.0 × 10−12; e 1.0 × 10−11; f 1.0 × 10−10; g 1.0 × 10−9. CC parameters were the same as in Fig. 3
Stability, regeneration and reproducibility of DNA biosensor
The stability of DNA biosensor was investigated by detecting 1.0 × 10−10 M target DNA every 24 h and recording the response value. When the modified electrode was not in use, it was stored in a refrigerator at 4 °C. The response values obtained were 7.83 × 10−5, 7.83 × 10−5, 7.79 × 10−5, 7.79 × 10−5, 7.79 × 10−5, 7.79 × 10−5, 7.76 × 10−5 C, respectively. The value of RSD obtained was 0.32 %, which indicates that this DNA biosensor possesses good stability.
The reproducibility of the DNA sensor was also investigated. In the experiment, we fabricated six DNA biosensors under the same conditions, and they were detected in 1.0 × 10−10 M complementary target DNA. The results are shown in Fig. S4 and indicate that the DNA sensor has good reproducibility.
The regeneration of the DNA sensor was performed by disassociating target DNA from the electrode by denaturing target DNA in hot water (80 °C) for 5 to 10 min, and it was then subjected to a second cycle of hybridization with the target DNA (1.0 × 10−10 M). The results show that the response of the biosensor obtained almost recovered to its original value (in five regenerations). The fifth regenerated sensor has 85.9 % response of the initial sensor (initial: 78 μC, final: 67 μC), indicating good reusability of the DNA biosensor.
Sample assay
We selected the human serum from a normal person to investigate the applicability of the biosensor. The treated blood serum was provided by Yiji Shan Hospital of Wuhu (the blood was treated with 4 % sodium citrate and centrifugation for 5 min at 10,000 rpm). 500 μL treated blood serum was added to 1.50 mL 10 mM Tris-HCl buffer (pH 7.4), then 2.0 μL 1.0 × 10−7 M or 2.0 × 10−8 M target DNA was added to the solution, and it was used as the hybridization solution. The analysis process is done according to the section of ‘Electrochemical detection of DNA hybridization’. The results are shown in Table 2. The recovery obtained was 87.10 % and 82.79 %, respectively.
Conclusions
In summary, we designed a sensitive DNA biosensor for sequence-specific DNA detection based on Au-NPs/PDA/MWCNTs film sensor interface. The Au-NPs/PDA/MWCNTs film significantly enhanced the performance of the electrochemical DNA biosensor. The DNA biosensor could detect as low as 3.5 fM target DNA and distinguish single-base mismatched target DNA under the optimal conditions. More important, the DNA biosensor has been applied in human serum samples for complementary target DNA analysis and provided satisfactory results.
References
Thaxton CS, Georganopoulou DG, Mirkin CA (2005) Gold nanoparticle probes for the detection of nucleic acid targets. Clin Chim Acta 363:120–126
Li HX, Rothberg LJ (2004) Label-free colorimetric detection of specific sequences in genomic DNA amplified by the polymerase chain reaction. J Am Chem Soc 126:10958–10961
Barhoumi A, Halas NJ (2010) Label-free detection of DNA hybridization using surface enhanced raman spectroscopy. J Am Chem Soc 132:12792–12793
Chen X, Lin YH, Lin LS, Chen GN, Yang HH (2011) A simple and ultrasensitive electrochemical DNA biosensor based on DNA concatamers. Chem Commun 47:12116–12118
He P, Dai LM (2004) Aligned carbon nanotube-DNA electrochemical sensors. Chem Commun 40:348–349
Wei MY, Guo LH, Famouri P (2011) DNA biosensors based on metallo-intercalator probes and electrocatalytic amplification. Microchim Acta 172:247–260
Zhou H, Liu J, Xu JJ, Chen HY (2011) Ultrasensitive DNA detection based on Au nanoparticles and isothermal circular double-assisted electrochemiluminescence signal amplification. Chem Commun 47:8358–8360
Liu G, Wan Y, Gau V, Zhang J, Wang LH, Song SP, Fan CH (2008) An enzyme-based E-DNA sensor for sequence-specific detection of femtomolar DNA Targets. J Am Chem Soc 130:6820–6825
Wang J (2003) Nanoparticle-based electrochemical DNA detection. Anal Chim Acta 500:247–257
Zhang YZ, Huang L (2012) Label-free electrochemical DNA biosensor based on a glassy carbon electrode modified with gold nanoparticles, polythionine, and grapheme. Microchim Acta 176:463–470
Larsson C, Rodahl M, Höök F (2003) Characterization of DNA immobilization and subsequent hybridization on a 2D arrangement of streptavidin on a biotin-modified lipid bilayer supported on SiO2. Anal Chem 75:5080–5087
Lucarelli F, Tombelli S, Minunni M, Marrazza G, Mascini M (2008) Electrochemical and piezoelectric DNA biosensors for hybridisation detection. Anal Chim Acta 609:139–159
Zhu NN, Chang Z, He PG, Fang YZ (2005) Electrochemical DNA biosensors based on platinum nanoparticles combined carbon nanotubes. Anal Chim Acta 545:21–26
Chang Z, Fan H, Zhao K, Chen M, He PG, Fang YZ (2007) Electrochemical DNA biosensors based on palladium nanoparticles combined with carbon nanotubes. Electroanalysis 2:131–136
Zhang SB, Wu ZS, Shen GL, Yu RQ (2009) A label-free strategy for SNP detection with high fidelity and sensitivity based on ligation-rolling circle amplification and intercalating of methylene blue. Biosens Bioelectron 24:3201–3207
Mao X, Jiang JH, Xu XM, Chu X, Luo Y, Shen GL, Yu RQ (2008) Enzymatic amplification detection of DNA based on “molecular beacon” biosensors. Biosens Bioelectron 23:1555–1561
Zhang J, Song SP, Zhang LY, Wang LH, Wu HP, Pan D, Fan CH (2007) A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat Protoc 2:2888–2895
Ren R, Leng CC, Zhang SS (2010) A chronocoulometric DNA sensor based on screen-printed electrode doped with ionic liquid and polyaniline nanotubes. Biosens Bioelectron 25:2089–2094
Hu KC, Lan DX, Li XM, Zhang SS (2008) Electrochemical DNA biosensor based on nanoporous gold electrode and multifunctional encoded DNA-Au bio bar codes. Anal Chem 80:9124–9130
Yang T, Zhang W, Du M, Jiao K (2008) A PDDA/poly(2,6-pyridinedicarboxylic acid)-CNTs composite film DNA electrochemical sensor and its application for the detection of specific sequences related to PAT gene and NOS gene. Talanta 75:987–994
Yang J, Yang T, Feng YY, Jiao K (2007) A DNA electrochemical sensor based on nanogold-modified poly-2,6-pyridinedicarboxylic acid film and detection of PAT gene fragment. Anal Biochem 365:24–30
Yang T, Zhou N, Zhang YC, Zhang W, Jiao K, Li GC (2009) Synergistically improved sensitivity for the detection of specific DNA sequences using polyaniline nanofibers and multi-walled carbon nanotubes composites. Biosens Bioelectron 24:2165–2170
Lei JP, Ju HX (2012) Signal amplification using functional nanomaterials for biosensing. Chem Soc Rev 41:2122–2134
Cosnier S, Holzinger M (2011) Electrosynthesized polymers for biosensing. Chem Soc Rev 40:2146–2156
Peng H, Zhang LJ, Soeller C, Travas-Sejdic J (2009) Conducting polymers for electrochemical DNA sensing. Biomaterials 30:2132–2148
Liu K, Wei WZ, Zeng JX, Liu XY, Gao YP (2006) Application of a novel electrosynthesized polydopamine-imprinted film to the capacitive sensing of nicotine. Anal Bioanal Chem 385:724–729
Wang GF, Huang H, Zhang G, Zhang XJ, Fang B, Wang L (2011) Dual Amplification strategy for the fabrication of highly sensitive interleukin-6 amperometric immunosensor based on poly-dopamine. Langmuir 27:1224–1231
Steel AB, Herne TM, Tarlov MJ (1998) Electrochemical quantitation of DNA immobilized on gold. Anal Chem 70:4670–4677
Zhang J, Song SP, Zhang LY, Wang LH, Wu HP, Pan D, Fan CH (2006) Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): Effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J Am Chem Soc 128:8575–8580
Li YL, Liu ML, Xiang CH, Xie QJ, Yao SZ (2006) Electrochemical quartz crystal microbalance study on growth and property of the polymer deposit at gold electrodes during oxidation of dopamine in aqueous solutions. Thin Solid Films 497:270–278
Wang J, Zhang SJ, Zhang YZ (2010) Fabrication of chronocoulometric DNA sensor based on gold nanoparticles/poly(L-lysine) modified glassy carbon electrode. Anal Biochem 396:304–309
Wang XQ, Zhang B, Lin XQ, Weng W (2011) Hybridization biosensor based on the covalent immobilization of probe DNA on chitosan-mutiwalled carbon nanotubes nanocomposite by using glutaraldehyde as an arm linker. Sensor Actuator B 156:599–605
Wei N, Chen JH, Zhang J, Wang K, Xu XW, Lin JH, Li GW, Lin XH, Chen YZ (2009) An electrochemical biosensor for detection of PML/RARA fusion gene using capture probe covalently immobilized onto poly-calcon carboxylic acid modified glassy carbon electrode. Talanta 78:1227–1234
Du P, Li HX, Mei ZH, Liu SF (2009) Electrochemical DNA biosensor for the detection of DNA hybridization with the amplification of Au nanoparticles and CdS nanoparticles. Bioelectrochemistry 75:37–43
Li F, Feng Y, Dong PJ, Yang LM, Tao B (2010) Gold nanoparticles modified electrode via simple electrografting of in situ generated mercaptophenyl diazonium cations for development of DNA electrochemical biosensor. Biosens Bioelectron 26:1947–1952
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 20675002) and the Natural Science Foundation of Anhui Province (No.1208085QB33).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 348 kb)
Rights and permissions
About this article
Cite this article
Dong, X., Lu, X., Zhang, K. et al. Chronocoulometric DNA biosensor based on a glassy carbon electrode modified with gold nanoparticles, poly(dopamine) and carbon nanotubes. Microchim Acta 180, 101–108 (2013). https://doi.org/10.1007/s00604-012-0900-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-012-0900-8