Abstract
A disposable electrochemical DNA-based biosensor was developed and applied as a screening device to detect an effect of a synthetically prepared quinazoline derivative on the surface-attached double stranded calf thymus DNA. Screen-printed carbon electrodes without and with multi-walled carbon nanotubes interface served as the signal transducer. The quinazoline interaction with DNA was investigated voltammetrically using DNA-bound electrochemical indicators such as [Co(phen)3]3+, [Ru(bpy)3]2+, methylene blue, the K3[Fe(CN)6] complex present in the solution phase as well as by electrochemical impedance spectroscopy. A severe damage to DNA at the incubation of the biosensor in quinazoline solution was found which leads to the loss of DNA from the electrode surface. Agarose gel electrophoresis was used to verify the results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The interactions of DNA with low molecular mass compounds are in centre of interest of the life sciences. Testing of such interactions using the DNA-based biosensors can help to evaluate health-risk chemicals and drugs including anticancer drugs, etc. [1]. The DNA biosensors are also widely used to detect the presence of pathogenic microorganisms, various types of damage to DNA, and protective effects of some substances towards the DNA structure [2]. A special feature of the DNA-based biosensors is in vitro the investigation of the surface attached DNA. DNA biosensor technologies are currently under intensive investigation owing to their great promise for the rapid and low cost investigation of potential drug agents [3].
In this work, we propose the electrochemical DNA-based biosensor as a screening device for the rapid test of potential antitumor agent interacting with the double helix of the nucleic acid. Concerning the surface-attached DNA voltammetric detection, various redox indicators can be used. The tris-(1,10-phenanthroline)cobalt(III) complex, [Co(phen)3]3+, interacts, depending on the ionic strength, with DNA through hydrophobic interactions with the interior of DNA and through electrostatic interactions with the negatively charged DNA backbone [4–6]. Tris-(2,2′-bipyridyl)ruthenium(II) complex, [Ru(bpy)3]2+, was used as the DNA intercalator already by Lerman in 1960s to define a molecule containing planar aromatic structure which inserts itself between the base pairs of double stranded DNA [7]. Recently, Lee et al. reported a voltammetric study on the interaction of this intercalator with DNA [8]. Catalytic oxidation of guanine residue by the Ru(III) complex provides a highly sensitive way to detect the DNA base damage [9]. The detection is based on the different accessibility and oxidation rate of guanine moiety in intact and damaged DNA [10].
Methylene blue (MB) is an aromatic organic dye that belongs to the phenothiazine family. The main interaction of MB with dsDNA is its intercalation between the base pairs of dsDNA [11], while other interactions such as a groove binding and electrostatic attraction also play some role which depends on the ionic strength and the MB/DNA molar ratio [12]. Compared to a label-free electrochemical method in detecting DNA damage, the utilization of MB shows the advantage in that its electrochemical reduction happens at the relatively low potential where the interferences from other electroactive species in a sample and background can be minimized [13, 14]. The function of the K3[Fe(CN)6] indicator present in solution phase is based on the electrostatic repulsion between the complex anion, [Fe(CN)6]3−, and the negatively charged DNA phosphate backbone. Cyclic voltammetry of [Fe(CN)6]3− was used as a routine method for the evaluation of the DNA presence at the electrode surface [15, 16].
Electrochemical impedance spectroscopy (EIS) is a powerful and effective tool for the investigation of electric features of the surface-modified electrodes in general [17]. With respect to advantages of the EIS detection of the surface attached DNA, particularly indicatorless procedures and commercially available equipments and software, impedimetric biosensors based on the conductive multi-walled carbon nanotubes (MWNT) and the DNA layers are also of great interest [18].
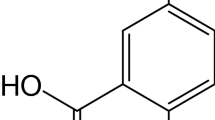
Quinazolines, the derivatives of 1,3-benzodiazine, are biologically active compounds used in pharmaceutical industry, agriculture and medicine. As we reported previously, many derivatives of quinazoline have antimicrobial [19], anticancer [20, 21] and DNA protective effects [22]. Various quinazolines exhibit also antiinflammatory, diuretic, anticonvulsant, antiallergic, antihypertensive and antiparkinsonian effects [23, 24]. In a preliminary study [25], the 3-(5-nitro-2-furyl)-9-chloro-5-morpholin-4-yl-[1,2,4]triazolo[4,3-c] quinazoline (Fig. 1) was shown as a compound with the strong effect on DNA. The aim of this paper is a detailed investigation of its reactivity towards surface-attached calf thymus dsDNA using the DNA-based biosensor and a variety of detection methods in order to obtain complex and verified results.
Experimental
Apparatus
Voltammetric measurements were performed with the Autolab apparatus and the software GPES (General Purpose Electrochemical System) version 4.9.005 from Eco Chemie, Utrecht (http://www.autolab-instruments.com). EIS measurements were carried out on the Autolab/FRA system with the potentiostat PGSTAT 12 and FRA-DSG, FRA-ADS modules, version 4.9.006 from Eco Chemie, Utrecht. Three-electrode assembly (Food Research Institute, Bratislava, http://www.vup.sk) consisting of working carbon electrode (25 mm2 geometric surface area), a silver/silver chloride reference electrode Ag/AgCl/SPCE (potential of 0.284 V vs conventional Ag/AgCl/saturated KCl electrode) and the same counter electrode was used. Electrophoretic experiments were performed with standard apparatus (Multigel G44, Biometra).
Preparation of DNA modified SPCE electrodes
DNA/SPCE
Five microliters of the DNA stock solution (0.1 mg ml−1) were dropped onto the bare SPE surface and allowed to evaporate to dryness over night.
DNA/MWNT/SPCE
Five microliters of the MWNT suspension (1 mg MWNT/2 ml in 1% sodium dodecyl sulphate) was dropped onto the bare SPCE surface and allowed to evaporate to dryness. MWNT/SPCE was then modified by dropping and evaporating 5 μl of the DNA stock solution (0.1 mg ml−1) over night.
Reagents
Calf thymus dsDNA was obtained from Merck, Darmstadt (http://www.merck.de). Its stock solution (0.1 mg ml−1) was prepared in 10 mM Tris–HCl and 1 mM EDTA solution of pH 8.0 and stored at −4 °C. Phosphate buffer solution of 1 mM, 5 mM and 100 mM (PBS), pH 7.0, was used for the electrochemical measurements.
Multi walled carbon nanotubes (OD 40–60 nm, ID 5–10 nm, length 0.5–500 μm) were obtained from Aldrich, Taufkirchen (http://www.sigma-aldrich.de). Their suspension was prepared in 1% aqueous solution of sodium dodecyl sulphate (SDS) from Sigma, Steinheim (http://www.sigma-aldrich.de).
The complex compound [Co(phen)3](ClO4)3 was synthesized in our laboratory according to [26] and checked by chemical analysis. The complex [Ru(bpy)3]Cl2 was synthesized at the Faculty of Natural Sciences, Charles University, Prague (http://www.natur.cuni.cz) according to [27] and checked by chemical analysis. MB was from Sigma, Steinheim and K3[Fe(CN)6] from Lachema, Brno. The quinazoline derivative was prepared by Špirková et al. [28]. Chromatographically pure quinazoline derivative was dissolved before use in DMSO (Biocom, Bratislava). The DMSO content in the cleavage medium never exceeded 0.4% (v/v).
Procedures
Before the measurement, the dry DNA modified electrode as an original biosensor was pretreated by immersion into the 1, 5 or 100 mM PBS, pH 7.0, for 5 min under stirring depending on the type of electrolyte used for the DNA detection. The electrochemical signals of selected DNA indicators were obtained as described below with optimum supporting electrolytes typically used in literature for the individual detection procedures. To detect an effect of the quinazoline derivative on DNA, the same sensor was incubated in the quinazoline solution (10, 50, 100 or 200 μg ml−1) in 5 mM PBS for a given time (10 min, 1 or 10 h) under stirring and rinsed with water. Then, the voltammetric response of the corresponding DNA indicator was obtained again in the appropriate supporting electrolyte.
Differential pulse voltammetry of [Co(phen)3]3+
The procedure described elsewhere [29] was used. Briefly, the DNA marker [Co(phen)3]3+ was accumulated from its 5 × 10−7 M solution in 5 mM PBS under stirring for 120 s at open circuit. The cathodic DP voltammogram was recorded immediately from 400 to −500 mV using pulse amplitude 100 mV, scan rate 10 mV s−1 and scan step 5 mV. The marker DPV peak current was evaluated against a base-line using the standard software and corrected to non-specific complex adsorption (I 0,DNA/SPCE − I 0,SPCE) to obtain the signal I 0. Then, the DNA modified electrode was regenerated by removing of the accumulated [Co(phen)3]3+ ions from the DNA layer by immersing the sensor into the solution of a high ionic strength (100 mM PBS) for 120 s under stirring. A negligible marker peak current was checked by the DPV record in the blank solution. After the incubation of the sensor in the quinazoline solution, rinsing with water and the 120 s marker accumulation from 5 × 10−7 M [Co(phen)3]3+ in 5 mM PBS, the DPV peak current was obtained in duplicate. The mean value (I) was calculated and corrected to non-specific complex adsorption (I DNA/SPCE − I SPCE). Finally, the normalized (relative) signal I/I 0 was received.
Square wave voltammetry of [Ru(bpy)3]2+
The procedure described elsewhere [30] was used. The sensor was immersed into 20 μM solution of [Ru(bpy)3]2+ in 1 mM PBS and the SWV anodic scan was recorded immediately from 300 to 1,500 mV using pulse amplitude 100 mV and scan rate 10 mV s−1. The peak current at 850 mV was against blank.
Cyclic voltammetry of MB
After an immersion of the biosensor into 20 μM MB in 1 mM PBS containing 20 mM NaCl, the cyclic voltammogram was recorded within the 0 to −700 mV potential range using the scan rate 50 mV s−1 and the cathodic peak current was evaluated against blank.
Cyclic voltammetry of K3[Fe(CN)6]
The cyclic voltammogram of 1 mM K3[Fe(CN)6] in 0.1 M PBS was recorded from 1,000 to −700 mV using the scan rate 50 mV s−1. The difference in the anodic to cathodic peak potential separation was evaluated.
Electrochemical impedance spectroscopy
The measurements were carried out in the presence of 1 mM K3[Fe(CN)6] as a redox probe in the 0.1 M PBS at the polarization potential 0 V in the frequency range 0.1 to 104 Hz and with the amplitude 10 mV.
Electrophoretic tests
Five microliters dsDNA (5 mg ml−1) were treated with quinazoline (10, 50, 100 or 200 μg ml−1) in 5 mM PBS at 22 °C for 10 min, 1 and 10 h. Then, the samples were subjected to electrophoresis at 40 V for 3 h in 2% (w/v) agarose gels complemented with ethidium bromide (1 mg l−1). Separated DNA fragments (DNA ladders) were visualized using a UV transilluminator (254 nm, Ultra-Lum Electronic UV Transilluminator, USA).
Results and discussion
Optimization of the DNA detection modes
Electrochemical response of the DNA marker [Co(phen)3]3+
The cathodic reduction of the complex was evaluated as the DNA signal. The DPV peak current obtained at a bare SPE immediately after its immersion into the [Co(phen)3]3+ solution represents about 5% and that after 120 s accumulation of the complex by simple adsorption (I SPE) about 15% of the signal measured after the 120 s accumulation of the complex (I DNA/SPE) at the DNA-modified SPCE (Fig. 2a). To evaluate the specific DNA response, the total peak current has to be corrected to the mean signal obtained at the bare SPCE (RSD = 10% for n = 10) by a subtraction of the last one (I = I DNA/SPCE − I SPCE).
a DPV curves of 5 × 10−7 M [Co(phen)3]3+ in 5 mM PBS, pH 7.0, obtained after 120 s accumulation at open circuit. Conditions: pulse amplitude 100 mV, scan rate 10 mV s−1, scan step 5 mV. b SWV scans of 20 μM [Ru(bpy)3]2+] in 1 mM PBS, pH 7.0 without accumulation. Conditions: pulse amplitude 100 mV, scan rate 10 mV s−1. c CV scans of 20 μM MB in 1 mM PBS containing 20 mM NaCl, pH 7.0 without accumulation. Conditions: scan rate 50 mV s−1. d CV scans of 1 mM K3[Fe(CN)6] in 0.1 M PBS pH 7.0, Conditions: scan rate 50 mVs−1
Electrochemical detection with [Ru(bpy)3]2+
The ruthenium complex [Ru(bpy)3]2+ was utilized as an electroactive DNA marker and catalyst of the guanine moiety oxidation (Fig. 2b). At the optimization of the procedure, the Ru(II) complex was accumulated at DNA/SPCE for 0, 30, 60, 90 or 120 s. Both, the specific (DNA/SPCE) and non-specific (SPCE) SWV signals have increased with time of accumulation, however, their ratio was approximately the same for individual time of accumulation. So, for the DNA detection, the procedure without accumulation was used. An effect of the PBS concentration on the [Ru(bpy)3]2+ signal was also tested and a significant influence of the ionic strength has on the SWV current was found (Fig. 3a). The Ru(II) signal increased with a decrease of the PBS concentration and 1 mM PBS was chosen for the measurement.
a The dependence of the 20 μM [Ru(bpy)3]2+ SWV peak current at the SPCE and DNA/SPCE on the concentration of PBS, pH 7.0. Conditions: amplitude 100 mV, scan rate 10 mV s−1. b The dependence of the 20 μM MB CV peak current at the SPCE and DNA/SPCE on the concentration of PBS, pH 7.0, containing 20 mM NaCl. Conditions: scan rate 50 mV s−1
Cyclic voltammetry of methylene blue (MB)
The reduction of MB was monitored by using CV. An adsorption of MB on DNA/SPCE leads to a significantly enhanced voltammetric response of MB comparing to bare SPCE (Fig. 2c). However, the signal did not increase with contact time (0, 30, 60, 90 and 120 s) of the DNA/SPCE with MB. An effect of the ionic strength on the interaction of MB with DNA was also investigated and a significant increase in the cathodic peak current with decreasing PBS concentration was found (Fig. 3b). For further experiments, 1 mM concentration of PBS containing 20 mM NaCl has been used.
Cyclic voltammetry of K3[Fe(CN)6]
The detection of DNA is based on a fact that the negatively charged [Fe(CN)6]3− complex anion in the solution phase can not reach the electrode covered with the polyanionic DNA due to a repulsive effect of the molecules. At the bare SPCE, the CV of K3[Fe(CN)6] in 0.1 M PBS is characterized by the anodic to cathodic peak potential separation of 510 mV. This ΔE p value is given by a relatively low reversibility of the redox couple at the carbon paste working electrode substrate. After covering the SPCE with DNA, however, the peak potential separation is even larger, about 880 mV and the CV peak currents are lower than those at bare SPCE (Fig. 2d). The difference in the peak potential separation was used to detect the presence of DNA layer at the electrode surface.
The effect of quinazoline
An effect of the quinazoline derivative on the DNA layer at the electrode surface has been evaluated after an incubation of the biosensor in quinazoline solution at 22 °C within the quinazoline concentration range from 10 to 200 μg ml−1. The voltammetric detection of damage to DNA has been accomplished using the electroactive compounds either bound to DNA (such as [Co(phen)3]3+, [Ru(bpy)3]2+, MB) or present in solution (the K3[Fe(CN)6] complex).
The quinazoline concentration dependent change of the surface-attached dsDNA was indicated by a decrease in the normalized (I/I 0) DPV signal of [Co(phen)3]3+ which represents a portion of DNA surviving the incubation in quinazoline (Fig. 4a). The quinazoline effect can be seen particularly after 1 and 10 h incubation. Similarly, the changes in the SWV signal of [Ru(bpy)3]2+ (Fig. 4b) and CVs of MB and K3[Fe(CN)6] were also found to increase with the quinazoline derivative concentration and incubation time (Fig. 4c,d).
The dependence of the DNA/SPCE response on the concentration of the quinazoline derivative. DNA/SPCE was incubated in the quinazoline in 5 mM PBS, pH 7.0, for 10 min (black bars), 1 h (white bars) and 10 h (grey bars) at 22 °C under stirring. Conditions: a normalized DPV peak current, I/I 0, of 5 × 10−7 M [Co(phen)3]3+ in 5 mM PBS, pH 7.0, 120 s accumulation, pulse amplitude 100 mV, scan rate 10 mV s−1, scan step 5 mV. b SWV peak current, I, of 20 μM [Ru(bpy)3]2+ in 1 mM PBS, pH 7.0, without accumulation, pulse amplitude 100 mV, scan rate 10 mV s−1. c CV cathodic peak current, I p, of 20 μM MB in 1 mM PBS, pH 7.0, without accumulation, scan rate 50 mV s−1. d CV anodic to cathodic peak potential separation, ΔE p, for 1 mM K3[Fe(CN)6] in 100 mM PBS, pH 7.0, scan rate 50 mV s−1
Concerning nature of the dsDNA change, the experiments with [Co(phen)3]3+, [Ru(bpy)3]2+ and MB indicate either a deep DNA structural degradation leading to DNA leaching from the electrode surface or a competitive adsorption of the quinazoline derivative on the DNA/SPCE. The tests with K3[Fe(CN)6] present in the solution phase showed the decrease in the peak potential separation after the DNA/SPCE treatment by the quinazoline derivative. Such ΔE p decrease, i.e. the increase in the electrochemical reversibility of the redox couple indicates the loss of negatively charged DNA from the electrode surface during the incubation and clearly confirms the damage to DNA and the potentiality of the quinazoline derivative under study to act as the DNA cleavage agent.
This observation was further confirmed by electrochemical impedance spectroscopy as the method for the detection of quality and changes at the electrode surface. For these experiments, the electrodes with nanostructured interface DNA/MWNT/SPCE were used because of higher impedimetric sensitivity of this DNA biosensor comparing to simple DNA/SPCE. The presence of MWNT on the SPCE electrode has significantly decreased its impedance, as one can expect, due to the high MWNT electrical conductivity. Typically, the system is simulated by a simple parallel combination of the capacitance (C) and the charge-transfer resistance (R p) in series with the supporting electrolyte resistance (R s). Moreover, the roughness exponent n [31] is used to express a deflection of the transducers surface from the theoretical homogenous or smooth surface where n = 1 [30]. At n values near to 1, the constant phase element should be reduced to capacitance C. The EIS detection window is given by a change in the R p value which is the highest in the presence of original dsDNA at the sensor surface and the smallest for the bare MWNT/SPCE without DNA.
The EIS parameters, particularly the polarization resistance (R p), have shown a significant change already after 1 and 10 h of incubation in the quinazoline solution (Table 1). The results correspond well with those obtained by using the DNA redox indicators and confirm the deep degradation of dsDNA in the presence of the quinazoline derivative.
Agarose gel electrophoresis was used as another conventional independent method to confirm the damage to DNA. The electrophoretic experiments have shown that at the given concentrations the quinazoline does not fission DNA in solution during 10 min incubation. However, at the cultivation of dsDNA with quinazoline for 1 and 10 h, the fission of dsDNA to small fragments takes place (Fig. 5). In the case of 10 h incubation, even a quinazoline concentration effect could be detected from the change of the picture in the lines 1 and 2 obtained for 10 and 50 μg ml−1 quinazoline, respectively. However, the sensitivity of the electrophoretic method to the quinazoline concentration changes here is lower than that of the DNA biosensors.
Two percent agarose gel electrophoresis of dsDNA previously treated with quinazoline in 5 mM phosphate buffer solution pH 7.0 for 10 min, 1 h and 10 h. Lines correspondence to quinazoline concentration: line 1 10 μg ml−1, line 2 50 μg ml−1, line 3 100 μg ml−1, line 4 200 μg ml−1, S standard, Cn negative control (non-treated dsDNA)
There are probably several types of quinazoline–DNA interactions. Consequences of these interactions involve changes in DNA. In many cases, changes in the structure of the DNA alter its thermodynamic stability which is manifested as a change in the functional properties of the DNA. We have found that the interaction of quinazoline derivative under study with DNA immobilized on the SPCE surface leads evidently to strand breaks, double helix opening and fragmentation of the attached DNA. To investigate the nature of the quinazoline–DNA interactions, more extensive study involving spectroscopic methods like IR, NMR and others would be necessary.
Conclusions
The search for new drugs, their evaluation and application requires an integrated action of many scientific disciplines such as pharmacy, biology, chemistry and medicine. An interest in the biosensor research is driven by increasing need of rather specific sensors for fast and routine tests in various fields of chemical analysis including the study of drugs and chemicals. The electrochemical DNA-based biosensors are known as experimentally simple devices which require only small amount of DNA. Using nanostructured materials, they efficiency can be significantly enhanced [32, 33]. The investigation of the electrochemical response of DNA before and after the interaction with a DNA-targeted drug can provide an evidence for its interaction mechanism.
In this paper, the good detection ability of such a biosensor at the study of deep changes of the dsDNA layer was demonstrated, particularly using instrumentally available combination of the K3[Fe(CN)6] cyclic voltammetry and electrochemical impedance spectroscopy. It was found that the quinazoline derivative causes the total DNA damage which depends on the concentration of quinazoline and incubation time. Generally, it is expected that the development through combined efforts in microelectronics, surface/interface chemistry, molecular biology and analytical chemistry will lead to further progress in the biosensor technology for the use in analytical biochemistry.
References
Del Carlo M, Di Marcello M, Perugini M, Ponzielli V, Sergi M, Mascini M, Compagnone D (2008) Electrochemical DNA biosensor for polycyclic aromatic hydrocarbon detection. Mikrochim Acta (in press). DOI 10.1007/s00604-008-0009-2
Lucarelli F, Kicela A, Palchetti I, Marrazza G, Mascini M (2002) Electrochemical DNA biosensor for analysis of wastewater samples. Bioelectrochemistry 58:113
Maciejewska D, Szpakowska I, Wolska I, Niemyjska M, Mascini M, Maj-Zurawska M (2006) DNA-based electrochemical biosensors for monitoring of bis-indoles as potential antitumoral agents, chemistry and x-ray crystallography. Bioelectrochemistry 69:1
Pang DW, Abruña HD (1998) Micromethod for investigation of the interactions between DNA and redox-active molecules. Anal Chem 70:3162
Labuda J, Bučková M, Vaníčková M, Mattusch J, Wennrich R (1999) Voltammetric detection of the DNA interaction with copper complex compounds and damage to DNA. Electroanalysis 11:101
Ovádeková R, Jantová S, Letašiová S, Štepánek I, Labuda J (2006) Nanostructured electrochemical DNA biosensors for the detection of berberine effect on DNA from cancer cells. Anal Bioanal Chem 386:2055
Lerman LS (1961) Structural considerations in the interaction of deoxyribonucleic acid and acridines. J Mol Biol 3:18
Lee JG, Yun K, Lim GS, Lee SE, Kim S, Park JK (2006) DNA biosensor based on the electrochemiluminescence of Ru(bpy)3 2+ with DNA-binding intercalators. Bioelectrochemistry 70:228
Johnston HD, Glasgow KC, Thorp HH (1995) Electrochemical measurement of the solvent accessibility of nucleobases using electron transfer between DNA and metal complexes. J Am Chem Soc 117:8933
Zhang Y, Hu N (2007) Cyclic voltammetric detection of chemical DNA damage induced by styrene oxide in natural dsDNA layer-by-layer films using methylene blue as electroactive probe. Electrochem Commun 9:35
Yang W, Ozsoz M, Hibbert DB, Gooding JJ (2002) Evidence for the direct interaction between methylene blue and guanine bases using DNA-modified carbon paste electrodes. Electroanalysis 14:1299
Kara P, Kerman K, Ozkan D, Meric B, Erdem A, Ozkan Z, Ozsoz M (2002) Electrochemical genosensor for the detection of interaction between methylene blue and DNA. Electrochem Commun 4:705
Kelly SO, Boon EM, Barton JK, Jackson NM, Hill MG (1999) Single-base mismatch detection based on charge transduction through DNA. Nucleic Acids Res 27:4830
Ostatná V, Dolinnaya N, Andreev S, Oretskaya T, Wang J, Hianik T (2005) The detection of deamination by electrocathalysis at DNA-modified electrodes. Bioelectrochemistry 67:205
Ferancová A, Ovádeková R, Vaníčková M, Šatka A, Viglaský R, Zima J, Barek J, Labuda J (2006) DNA modified screen-printed electrodes with nanostructured films of multi-wall carbon nanotubes, hydroxyapatite and montmorillonite. Electroanalysis 18:163
Maeda M, Mitsuhashi Y, Nakano K, Takagi M (1992) DNA-immobilized gold electrode for DNA-binding drug sensor. Anal Sci 8:83
Guan JG, Miao YQ, Zhang QJ (2004) Review: impedimetric biosensors. J Biosci Bioeng 97:219
Guo S, Qu X, Dong S (2007) Nanoelectrode ensembles based on semi-interpenetrating network of carbon nanotubes. Electrochim Acta 52:6186
Jantová S, Ovádeková R, Letašiová S, Špirková K, Stankovská Š (2005) Antimicrobial activity of some substituted triazoloquinazolines. Folia Microbiol 50:90
Jantová S, Letašiová S, Repický A, Ovádeková R, Lakatoš B (2006) The effect of 3-(5-nitro-2-thienyl)-9-chloro-5-morpholin-4-y1[1,2,4]triazolo [4,3-c]quinazoline on cell growth, cell cycle, induction of DNA fragmentation, and activity of caspase 3 in murine leukemia L1210 cells and fibroblast NIH-3T3 cells. Cell Biochem Funct 24:519
Teraishi F, Kagawa S, Watanabe T, Tango Y, Kawashima T, Umeoka T, Nisizaki M, Tanaka N, Fujiwara T (2005) ZD1839 (Gefitinib, ‘Iressa’), an epidermal growth factor receptor-tyrosine kinase inhibitor, enhances the anti-cancer effects of TRAIL in human esophageal squamous cell carcinoma. FEBS Lett 579:4069
Ovádeková R, Jantová S, Labuda J (2005) Detection of the effective DNA protection by quinazolines using a DNA-based electrochemical biosensor. Anal Lett 38:2625
Marti M, Trapella C, Viaro R, Morari M (2007) The nociceptin/orphanin FQ receptor antagonist J-113397 and L-DOPA additively attenuate experimental parkinsonism through overinhibition of the nigrothalamic pathway. J Neurosci 27:1297
Chao Q, Deng L, Shih H, Leoni LM, Genini D, Carson DA, Cottam HB (1999) Substituted isoquinolines and quinazolines as potential antiinflammatory agents. Synthesis and biological evaluation of inhibitors of tumor necrosis factor alpha. J Med Chem 42:3860
Jantová S, Letašiová S, Ovádeková R, Mučková M (2006) Cytotoxic/zntoproliferative effects of new [1,2,4]triazolo[4,3-c] quinazolines in tumor cell lines HeLa and B16. Neoplasma 53:291
Dollimore LS, Gillard RD (1973) Optically active co-ordination compounds. Part XXXII. Potassium (+)tris-[l-cysteinesulphinato(2−)-SN]cobaltate(III): a versatile agent for resolution of 3+ species. J Chem Soc 78:933
Juany Q, Sun S, Nakansson M, Langel K, Ylinen T, Sukni J, Kulhala S (2006) Electrochemiluminescence and chemiluminescence of a carboxylic acid derivative of ruthenium(II) tris-(2,2′-bipyridine) chelate synthesized for labeling purposes. J Lumin 118:265
Špirková K, Stankovský Š (1991) Amidinoyl isothiocyanates in the synthesis of condensed quinazolines. Preparation of 3-aryl, 5,9-disubstituted-s-triazolo[4,3-c] quinazolines. Collect Czechoslov Chem Commun 56:1719
Labuda J, Bučková M, Heilerová Ľ, Šilhár S, Štepánek I (2003) Evaluation of redox properties and anti-/prooxidant effects of selected flavonoids by DNA-based electrochemical biosensor. Anal Bioanal Chem 376:168
Oliveira-Brett AM, Silva LA, Farace G, Vadgama P, Brett MACh (2003) Voltammetric and impedance studies of inosine-5′-monophosphate and hypoxanthine. Bioelectrochemistry 59:49
Hays CWH, Millner PA, Prodromidis MI (2006) Development of capacitance based immunosensors on mixed self-assembled monolayers. Sens Actuators B Chem 114:1064
Merkoçi A (2006) Carbon nanotubes: exciting new materials for microanalysis and sensing. Mikrochim Acta 152:155
Liu Y, Hu LM, Yang SQ (2008) Amplification of bioelectrocatalytic signalling based on silver nanoparticles and DNA-derived horseradish peroxidase biosensors. Mikrochim Acta 160:357, DOI 10.1007/s00604-007-0817-9
Acknowledgements
This work was supported by the Research and Development Assistance Agency under contract no. APVT-20-015904 and by the Ministry of Education (Applied Research Project AV/4/0103/06). The authors thank Assoc. Professor Katarína Špirková and Assoc. Professor Štefan Stankovský, STU, for the sample of quinazoline derivative, Professor Jiří Barek, Charles University, Prague, for the [Ru(bpy)3]Cl2 complex preparative as well as Dr. Guzel Ziyatdinova, Kazan State University, Russia, for the technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Labuda, J., Ovádeková, R. & Galandová, J. DNA-based biosensor for the detection of strong damage to DNA by the quinazoline derivative as a potential anticancer agent. Microchim Acta 164, 371–377 (2009). https://doi.org/10.1007/s00604-008-0068-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-008-0068-4