Abstract
Aims
The study aimed to screen the HNF1A and HNF4A mutation in a large Chinese cohort of high clinical suspicion of maturity-onset diabetes of the young (MODY) patients and characterize the clinical features of those patients. The performance of hsCRP as a biomarker to differentiate MODY3 from early onset T2DM was also evaluated.
Methods
A total of 74 patients with a strong clinical suspicion of MODY from 59 families and 33 newly diagnosed early-onset T2DM were included. HNF1A and HNF4A mutations were analyzed by Sanger sequencing. ROC curves were used to identify the optimal cutoff of hsCRP.
Results
One novel (c.864_865insG) and six recurrent HNF1A mutations (R203H, R263H, P379T, L422P, P519L and c.873delC) in 17 patients from 8 families (13.6%), as well as one novel HNF4A (R331H) mutation were identified. Nonspecific clinical presentations were observed in MODYX compared to MODY3 patients. MODY3 subjects exhibited with younger, lower BMI, TG, fasting and postprandial C-peptide, higher HDL than T2DM. Particularly, we confirmed serum hsCRP was lower in MODY3 than T2DM. ROC curve showed a good discrimination with an AUC of 0.852 and identified a cutoff hsCRP of 0.79 (75% sensitivity and 83% specificity). Good glycemic control was observed in all identified patients after switching to glimepiride therapy.
Conclusions
The prevalence of HNF1A mutation was relatively lower in Mainland China and HNF4A mutation was rare. Serum hsCRP concentrations performed well in discriminating MODY3 from T2DM. Molecular diagnosis of MODY3/1 did transform management in clinical practice and facilitated the glycemic control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maturity-onset diabetes of the young (MODY) is a clinically and genetically heterogeneous monogenic disorders characterized by autosomal dominant inheritance, early onset and pancreatic beta cells dysfunction [1]. Presently, at least 14 causative genes involved in insulin secretion have been identified for MODY. Mutation in genes encoding glucokinase (MODY2), hepatic nuclear factors-1A (MODY3) and HNF4A (MODY1) are the three most common subtypes, accounting for more than 90% MODY patients [2], but the relative prevalence varied considerably among different ethnicity [3]. Small studies from Hong Kong reported that HNF1A mutation was responsible for only 5–10% MODY and no MODY1 was identified in Chinese [4,5,6], compared to 52% and 10% MODY patients with HNF1A and HNF4A mutation, respectively, in UK [7]. However, the prevalence of MODY3 and MODY1 in Mainland China remains unknown.
In contrast with MODY2 characterized by mild fasting hyperglycemia and the absence of complications, patients with HNF1A or HNF4A mutation demonstrate with progressive beta cell failure and high risk of microvascular and macrovascular complications due to increasing hyperglycemia [8]. Another significantly clinical implication is that afflicted cases are extremely sensitive to sulfonylureas [9]. Thus, a definitive molecular diagnosis facilitates optimal treatment and good glycemic control. However, hitherto there were fewer reports on MODY1 and MODY3 families in Chinese population. It is noteworthy that diabetes is dramatically prevalent in China, where the number of diabetes ranks first throughout the world [10]. This clearly suggests that the vast majority of MODY3 and MODY1 individuals are currently undiagnosed. Limited clinicians experience and restrictive selection criteria for genetic testing may partly explain the low referral rate in Chinese population. In recent years, an older age of onset had been frequently observed in HNF1A mutation carriers [11]. Hence, an extensive genetic screening in the higher age threshold is warrant.
On the other hand, since the prevalence of young-onset diabetes and overweight is increasing [10], it becomes more challenging for clinicians to differentiate HNF1A/4A MODY from other forms of diabetes owing to the significantly overlapping clinical features. Although the correct diagnosis depends on molecular genetic testing, it is too expensive and not available in many hospitals. Consequently, a cost effective and practical biomarker that helps to pick up patients with high probability of HNF1A/4A mutation for genetic screening is desirable. A body of evidence shows that high sensitivity C reaction protein (hsCRP) is significantly lower in MODY3 patients than other types of diabetes, and hsCRP discriminates MODY3 well from young-onset type 2 diabetes mellitus (T2DM) [12, 13]. However, the diagnostic value of hsCRP in Chinese has not been investigated yet.
Combine with these, this study aimed to first screen the HNF1A and HNF4A mutation in a large Chinese cohort of high clinical suspicion of MODY3 or MODY1 patients using a wider selection criterion (up to 45 yeas) to determine the prevalence of MODY3/1 in Mainland China, and analyzed the clinical features of those patients. In addition, the study evaluated the clinical validity of hsCRP as a diagnostic marker to differentiate HNF1A mutation carriers from early onset T2DM.
Materials and methods
Subjects
A total of 74 patients with a strong clinical suspicion of MODY3 or MODY1 from 59 families were studied. These subjects were consecutively recruited from the outpatient clinic of Endocrinology at Peking Union Medical College Hospital (PUMCH), Beijing, China, between January 2014 and December 2016. All proband met the following criteria: (1) the age at diagnosis of diabetes ≤ 45 year; (2) family history of diabetes in at least two generations with autosomal dominant mode of inheritance; (3) the absence of pancreatic islet autoantibodies including glutamic acid decarboxylase antibody (GAD), protein tyrosine phosphatase antibody (IA2) and islet cell antibody (ICA); (4) no marked obesity or evidence of insulin resistance. We excluded patients with a renal disorder, persistently mild fasting hyperglycemia, a maternally inherited diabetes and deafness. In addition, 33 unrelated Chinese with newly identified early-onset T2DM aged less than 45 years were recruited as a control group. These patients were diagnosed with diabetes within 1 year. Subjects with hsCRP > 10 mg/L were removed.
Genetic analysis
All these patients were sequentially underwent HNF1A and HNF4A gene mutation analysis. Genomic DNA was extracted from peripheral blood lymphocytes using a DNA extraction kit (Qiagen, Germany). All exons, the intron–exon boundaries and the promoter regions of HNF1A and HNF4A gene were amplified by polymerase chain reaction (PCR). The primers were available on request. Sequencing of purified PCR products in both forward and reverse directions was performed on an ABI3730 automated sequencer (Applied Biosystems. Foster City. CA. USA) using the chain-termination method. The NCBI BLAST database was applied to identify variants by aligning with reference sequences NM_000545.6 (HNF1A) and NM_000457.4 (HNF4A). Then, three databases including Exome Variant Sever (http://evs.gs.washington.edu/EVS), dbSNP database in NCBI (http://www.ncbi.nlm.nih.gov/snp/) and UCSC genome bank were searched to exclude single nucleotide polymorphisms (SNPs). Furthermore, the identified variants should not been found in 100 healthy control. For putative novel missense mutation, SIFT (http://sift.jcvl.org), PolyPhen2 (http://genetics.bwh.harvard.edu/pph2) and Mutation T@sting (http://www.muatationtaster.org) software were used to predict the pathogenicity.
Clinical and laboratory examinations
Clinical parameters including age at diagnosis of diabetes, height, weight, family history of diabetes, past medical history, treatment before referring to PUMCH and physical examination were recorded for each participant. Body mass index (BMI) was calculated as weight (kg) divided by the square of height (m2). Routine biochemical examinations included measurement of live and renal function, fasting and postprandial glucose, serum lipid profiles and hsCRP with an automatic biochemical analyzer (AU5800; Beckman Coulter, USA). The serum insulin and C-peptide levels were measured by direct chemiluminescence immunoassay (SIMENS ADVIA Centaur XP, Germany). The high performance liquid chromatography method was used to determine glycated hemoglobin (HbA1c) levels. GAD and IA2 were determined by ELISA (SIMENS ADVIA Centaur XP, Germany), and ICA were measured using indirect immunofluorescence (SIMENS ADVIA Centaur XP, Germany).
Statistical analysis
All the statistical analyses were performed using SPSS version 16.0 and Graphpad Prism 6.0. The normal distribution of continuous variables was evaluated by Kolmogorov–Smirnov test. Comparisons between two groups (MODY group vs MODY negative group or MODY group vs T2DM group) were conducted using Student’s t test for normally distributed continuous variables and non-parametric test for skewed continuous variables expressed as median and range. A Chi-square test was used for categorical variables expressed as percentage. Receiver operating characteristic (ROC) curve analysis was applied to evaluate the performance of hsCRP in distinguishing MODY3 from T2DM, and also used to identify cutoffs of hsCRP with optimal sensitivity and specificity. P < 0.05 was considered significant.
Results
Genetic screening of HNF1A and HNF4A mutation
HNF1A gene mutation analysis was conducted in all subjects diagnosed clinically as MODY. In total, seven different mutations in 17 patients from 8 families were identified, with a mutation pick-up rate of 13.6%. Among these mutations, five were missense, one insertion and one deletion mutation. The insertion mutation (c.864_865insG) had not been reported previously. Five mutations (c.864_865insG, c.873delC, c.1135 C > A, c.1265 T > C and c.1556 C > T) were located in transactivation domain and two (c.608G > A and c.788G > A) in the DNA-binding domain. Each mutation was identified only in a single family except for c.873delC. Three out of seven mutations (42.9%) were resided in exon 4. The detailed information of HNF1A mutations were shown in Table 1 and Fig. 1.
For MODY individuals with no HNF1A gene mutation, HNF4A mutation testing was then performed. Only one HNF4A mutation was detected in two patients from one family (Fig. 2). The MODY1 subjects were heterozygous for a novel G–A transversion at position 992 (c.992 G > A) resulting in a replacement of arginine by histidine at amino acid 331 (p.R331H). The Polyphen-2 bioinformatics tool predicted the R331H substitution to be probably damaging, “Disease causing” and “Intolerant” was showed by Mutation Taster and SIFT, respectively. Furthermore, R331 residue was highly conserved across ten different species (Figure S1). These results indicated R331H mutation was pathogenic.
Clinical characteristics of MODY3 subjects
For MODY3 patients, the mean age of diagnosis of diabetes was 25 years, but the onset of diabetes above 35 years old was found in five subjects. Fourteen patients were misdiagnosed other types of diabetes more than one year, and the longest length was 16 years. BMI of all MODY3 patients was below 24 kg/m2. Compared to non-HNF1A mutation carriers, no significantly statistical differences in glucose and lipid metabolism traits were observed except for lower postprandial C-peptide (p = 0.029) (Table 2).
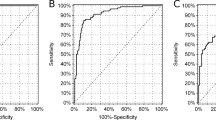
Besides, MODY3 patients were younger, leaner and exhibited with lower fasting and postprandial C-peptide compared to T2DM (p < 0.001). Serum concentrations of triglycerides (TG) were lower (p = 0.006), whereas high-density lipoprotein cholesterol (HDL-c) levels were higher in MODY3 than T2DM subjects (p = 0.004). Particularly, the levels of hsCRP were significantly decreased in MODY3 patients [0.17(0.07,0.73)] in comparison with T2DM [2.27(0.68,4.38); (p < 0.001)] (Table 2; Fig. 3A).
Clinical presentation of MODY1 patients
The two identified MODY1 patients were from one family, that is proband and his mother. The proband, a 10-year-old boy, was born at term with a birth weight of 4.4 kg. He was the only child of a non-consanguineous couple. At 8 years, he presented with polyuria, but paid no attention. High postprandial serum glucose (13.1 mmol/L) was ever observed unintentionally at home by detection capillary blood glucose, but no treatment was received. Two years later, he was referred to our hospital for the first time for further examination and management. His height was 147.5 cm (+ 1 to + 2SD) and weight was 46 kg. Liver and renal function was normal. HbA1c was 8.6% (70 mmol/mol). Relatively decreased pancreatic islet function with fasting C-peptide of 0.88 μg/L and postprandial C-peptide of 2.15ug/L was found. Elevated LDL-c (2.89 mmol/L) was measured. The levels of TG and hsCRP were within normal range and the value was 0.65 mmol/L and 0.95 mg/L, respectively. His mother had a history of gestational diabetes mellitus. Four years after delivery, hyperglycemia was detected because of ketosis and then insulin treatment was initiated. Unfortunately, her laboratory examination was not available.
Diagnostic value of hsCRP
ROC curve analysis was used to further evaluate the performance of hsCRP as a diagnostic marker to discriminate MODY3 patients from newly diagnosed early onset T2DM. The results showed good discrimination with an area under the curve (AUC) of 0.852 and identified a cut-off hsCRP < 0.79 mg/L with 75% sensitivity and 83% specificity (Fig. 3b).
Efficacy of sulfonylureas treatment
Among the 17 MODY3 patients, only two patients received correct treatment with sulfonylureas at the onset of diabetes because of identification HNF1A mutation in our hospital. Insulin treatment was initially given in more than half of MODY3 patients. 11.8% patients were treated with insulin and oral hypoglycemic agents, 29.4% other oral hypoglycemic medicine only. After molecular diagnosis with MODY3, insulin treatment as well as other oral hypoglycemic agents was gradually off and switched to glimepiride. HbA1c levels decreased markedly to below 7% (53 mmol/mol) in most patients (Fig. 4) following 3 months of glimepiride treatment and no adverse effect was observed. The MODY1 proband was also transferred from insulin to glimepiride successfully. HbA1c was dropped to 6.2% (44 mmol/mol) after 6 months of this transition. During the follow-up period, no deterioration in glycemic control was observed in all MODY1/3 patients.
Discussion
To the best of our knowledge, this was the first study in Mainland China to screen the HNF1A and HNF4A mutation in a large cohort of Chinese MODY patients. We identified one novel and six recurrent HNF1A gene mutations, and one novel missense mutation in HNF4A gene. The prevalence of MODY3 and MODY1 was 13.6% and 1.7%, respectively. In addition, we confirmed that hsCRP levels can be used as a biomarker to help distinguish MODY3 from T2DM.
In Chinese population with an epidemic of diabetes, there are only three reports from HongKong focusing on the frequency of HNF1A and HNF4A mutation, in which prevalence of 5–10% for MODY3 and no MODY1 was reported [4,5,6]. In this study, extended criteria of age of diabetes onset to 45 increased pick up rate from 11.8 to 13.6%. The pick up rate was similar to Italy (14%) [14] but higher than US (4.4%) [15]. Unlikely MODY2, MODY3 patients usually developed diabetes after puberty, and the penetrance increased with age, which was almost complete by 55 years [16]. This was consistent with our findings that five MODY3 patients were diagnosed with diabetes after the age of 35 years, and even one onset at 51 years old. Thus, the higher age threshold for screening HNF1A mutation was necessary. On the other hand, excluding individuals with phenotype resembling MODY2 might also contribute to improving the positive mutation rates. However, the prevalence of MODY3 in Chinese population was strikingly lower than what had been reported in UK (52%) [7], Germany (31%) [17], Norway (29%) [18], Spain (35%) [19] and Denmark (36%) [20]. This discrepancy was more likely explained by different recruitment criteria for genetic test and ethnicity background.
In contrast with MODY2 and MODY3, MODY1 was relatively uncommon [2]. Till now, less than ten MODY1 patients were described in China. In the present study, we identified only one HNF4A mutation. The novel missense mutation c.992G > A occurred at amino acid position 331 of exon 8 in transactivation domain, where an arginine was replaced by histidine. The Arg331 code was highly conserved across various species, and the mutation was absent in 100 health controls. Furthermore, three bioinformatics tools we applied predicted R331H mutation to be pathogenic variants, and the mutation cosegregated with the phenotype in the pedigree. Accordingly, this mutation was responsible for the typical clinical features of the index case. In line with previous description, the proband showed macrosomia and increased LDL-c levels. It is worth mentioning that the birth weight of GCK-deficient fetuses varied depending on different GCK mutations and whether their mothers were affected. Consequently, a close fetal growth monitoring through frequent ultrasounds is valuable in the management of MODY2 pregnancies [21].
Although HNF1A mutations were widely distributed in different exons, of which 37.5% (3 out of 8 families) occurred in a polyC-tract of exon4, suggesting it was vulnerable to mutation and still a hotspot mutation in Chinese. This was consistent with previous observations in other population [22]. The novel insertion mutation c.864_865insG at code 288 resulted in a frameshift generating an early termination signal at amino acid 316. The truncated protein lacking C-terminal transactivation domain was expected to severely impair transcriptional activity. With regard to subjects carrying different types or location of HNF1A mutation, no genotype and phenotype correlations were observed. Even the same HNF1A mutation carriers, the penetrance and expression varied. Other genetic or environmental factors might influence phenotype as well.
For most included MODY patients, both HNF1A and HNF4A mutation were not detected. While GCK and HNF1B mutations were not analyzed in this study, we excluded subjects whose clinical features were strongly suggestive of MODY2 or MODY5. Consequently, common MODY subtypes were not the major causes of these MODY patients. Whole exome sequencing was needed to further investigate other rare or new MODY genes in future. To reflect the real-life situations encountered by clinicians, we compared MODY3 patients with non-MODY3/1 patients, and nonspecific clinical characteristic was observed. Thus, to select MODY patients for HNF1A mutation analysis only depending on clinical manifestations remained challenging for clinicians.
Despite an accurate molecular diagnosis of MODY3 was crucial for optimizing management, a vast majority of MODY3 patients were frequently misclassified as T2DM or other type diabetes [23]. In this study, 84.2% identified patients were misdiagnosed for more than 1 year. Therefore, a cheap and widely available biomarker was considerably valuable in assisting selection of individuals for HNF1A sequencing. The promoter of CRP gene harbored binding sites of transcription factors hnf1a which directly regulated CRP expression [24]. Loss of function mutations in HNF1A gene responsible for MODY3 had been shown to confer a substantial CRP reduction [12, 13]. Our study confirmed that the levels of serum hsCRP were significantly lower in MODY3 than T2DM patients, and ROC curve showed a good discrimination with a AUC of 0.852 and identified a cutoff hsCRP of 0.79 (75% sensitivity and 83% specificity). Interestingly, the results were consistent with reports from UK, which indicated that hsCRP distinguished MODY3 from T2DM with hsCRP < 0.75 mg/L showing 79% sensitivity and 70% specificity (AUC = 0.84) [25]. These evidence suggested that hsCRP was a clinically valid biomarker used to determine whether HNF1A mutation testing was initiated. Interestingly, Delvecchio M recently proposed to screen HNF1A gene first in patients with primarily fasting plasma glucose ≤ 150 mg/dl and HbA1c > 7.3%, irrespective of other clinical information. Large sample studies are warranted for further evaluation [26].
The clinical benefits of molecular diagnosis MODY3 and MODY1 were that these types of diabetes were classically sensitive to sulfonylurea treatment. A randomized crossover trial indicated that MODY3 patients had a 5.2-fold greater response to gliclazide than to metformin [27]. In this patient cohort, MODY3 and MODY1 patients were properly offered low-dose glimepiride therapy following genetic diagnosis. Insulin therapy was safely stopped and patients reported that their quality of life was improved. For all patients, blood glucose was controlled well throughout the follow-up period. Previous observational study also found most MODY3 patients transferred off insulin to sulfonylurea successfully and the efficacy appeared to be sustained over 3 years [28], similar response was also seen in MODY1 patients [29]. Importantly, due to the progressive nature of beta cell function, these patients should be closely monitored during long-term follow-up.
Some limitations should be stated. First, Sanger sequencing utilized in this study could not capture large deletions, which might underestimate the positive rate of HNF1A and HNF4A mutation. The method of multiplex-ligation dependent probe amplification (MLPA) should be applied in future. Second, a small number of MODY3 and MODY1 subjects restricted the analysis of genotype and phenotype correlations. Third, certain drugs such as statins were believed to reduce the CRP level. Nevertheless, the drug history of some cases with T2DM was not available, thus we could not evaluate its influence on our results.
Conclusions
The prevalence of HNF1A mutation was relatively lower in Mainland China and HNF4A mutation was rare, while the misdiagnosis rate was high. We identified one novel HNF1A and HNF4A mutation, respectively, which were probably pathogenic and expand the mutation spectrum of the two genes. The etiology of large number of non-MODY1/3 subjects required a broader range of investigation for possible pathogenic genes in future. Molecular diagnosis of MODY3 and MODY1 did transform management in clinical practice and facilitated the glycemic control. In addition, serum hsCRP concentrations were lower in MODY3 than T2DM and performed well in discriminating MODY3 from T2DM. hsCRP could be a useful biomarker in clinical practice to assist selecting patients for diagnostic HNF1A genetic testing.
References
Velho G, Froguel P (1998) Genetic, metabolic and clinical characteristics of maturity onset diabetes of the young. Eur J Endocrinol 138:233–239
Gardner DS, Tai ES (2012) Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab Syndr Obes 5:101–108
Kleinberger JW, Pollin TI (2015) Undiagnosed MODY: time for action. Curr Diab Rep 15:110
Ng MC, Lee SC, Ko GT, Li JK, So WY, Hashim Y et al (2001) Familial early-onset type 2 diabetes in Chinese patients: obesity and genetics have more significant roles than autoimmunity. Diabetes Care 24:663–671
Xu JY, Chan V, Zhang WY, Wat NM, Lam KS (2002) Mutations in the hepatocyte nuclear factor-1alpha gene in Chinese MODY families: prevalence and functional analysis. Diabetologia 45:744–746
Xu JY, Dan QH, Chan V, Wat NM, Tam S, Tiu SC et al (2005) Genetic and clinical characteristics of maturity-onset diabetes of the young in Chinese patients. Eur J Hum Genet 13:422–427
Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S (2010) Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 53:2504–2508
Isomaa B, Henricsson M, Lehto M, Forsblom C, Karanko S, Sarelin L (1998) Chronic diabetic complications in patients with MODY3 diabetes. Diabetologia 41:467–473
Sovik O, Njolstad P, Folling I, Sagen J, Cockburn BN, Bell GI (1998) Hyperexcitability to sulphonylurea in MODY3. Diabetologia 41:607–608
Hu C, Jia W (2018) Diabetes in China: epidemiology and genetic risk factors and their clinical utility in personalized medication. Diabetes 67:3–11
Timsit J, Saint-Martin C, Dubois-Laforgue D, Bellanne-Chantelot C (2016) Searching for maturity-onset diabetes of the young (MODY): when and what for? Can J Diabetes 40:455–461
Thanabalasingham G, Shah N, Vaxillaire M, Hansen T, Tuomi T, Gasperikova D et al (2011) A large multi-centre European study validates high-sensitivity C-reactive protein (hsCRP) as a clinical biomarker for the diagnosis of diabetes subtypes. Diabetologia 54:2801–2810
Owen KR, Thanabalasingham G, James TJ, Karpe F, Farmer AJ, McCarthy MI et al (2010) Assessment of high-sensitivity C-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1A mutations. Diabetes Care 33:1919–1924
Gragnoli C, Cockburn BN, Chiaramonte F, Gorini A, Marietti G, Marozzi G et al (2001) Early-onset type II diabetes mellitus in Italian families due to mutations in the genes encoding hepatic nuclear factor 1 alpha and glucokinase. Diabetologia 44:1326–1329
Pihoker C, Gilliam LK, Ellard S, Dabelea D, Davis C, Dolan LM et al (2013) Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for diabetes in youth. J Clin Endocrinol Metab 98:4055–4062
Harries LW, Ellard S, Stride A, Morgan NG, Hattersley AT (2006) Isomers of the TCF1 gene encoding hepatocyte nuclear factor-1 alpha show differential expression in the pancreas and define the relationship between mutation position and clinical phenotype in monogenic diabetes. Hum Mol Genet 15:2216–2224
Schober E, Rami B, Grabert M, Thon A, Kapellen T, Reinehr T et al (2009) Phenotypical aspects of maturity-onset diabetes of the young (MODY diabetes) in comparison with type 2 diabetes mellitus (T2DM) in children and adolescents: experience from a large multicentre database. Diabet Med 26:466–473
Bjørkhaug L, Sagen JV, Thorsby P, Søvik O, Molven A, Njølstad PR (2003) Hepatocyte nuclear factor-1 alpha gene mutations and diabetes in Norway. J Clin Endocrinol Metab 88:920–931
Costa A, Bescós M, Velho G, Chêvre J, Vidal J, Sesmilo G et al (2000) Genetic and clinical characterisation of maturity-onset diabetes of the young in Spanish families. Eur J Endocrinol 142:380–386
Johansen A, Ek J, Mortensen HB, Pedersen O, Hansen T (2005) Half of clinically defined maturity-onset diabetes of the young patients in Denmark do not have mutations in HNF4A, GCK, and TCF1. J Clin Endocrinol Metab 90:4607–4614
Bitterman O, Tinto N, Franzese A, Iafusco F, Festa C, Mozzillo E et al (2018) Glucokinase deficit and birthweight:does maternal hyperglycemia always meet fetal needs? Acta Diabetol. https://doi.org/10.1007/s00592-018-1198-8
Ellard S, Colclough K (2006) Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1 alpha (HNF1A) and 4 alpha (HNF4A) in maturity-onset diabetes of the young. Hum Mutat 27:854–869
Thanabalasingham G, Owen KR (2011) Diagnosis and management of maturity onset diabetes of the young (MODY). BMJ 343:d6044
Nishikawa T, Hagihara K, Serada S, Isobe T, Matsumura A, Song J et al (2008) Transcriptional complex formation of c-Fos, STAT3, and hepatocyte NF-1 alpha is essential for cytokine-driven C-reactive protein gene expression. J Immunol 180:3492–3501
McDonald TJ, Shields BM, Lawry J, Owen KR, Gloyn AL, Ellard S et al (2011) High-sensitivity CRP discriminates HNF1A-MODY from other subtypes of diabetes. Diabetes Care 34:1860–1862
Delvecchio M, Salzano G, Bonura C, Cauvin V, Cherubini V, d’Annunzio G et al (2018) Can HbA1c combined with fasting plasma glucose help to assess priority for GCK-MODY vs HNF1A-MODY genetic testing? Acta Diabetol 55:981–983
Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT (2003) Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362:1275–1281
Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT (2009) A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med 26:437–441
Pearson ER, Pruhova S, Tack CJ, Johansen A, Castleden HA, Lumb PJ et al (2005) Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4alpha mutations in a large European collection. Diabetologia 48:878–885
Acknowledgements
The study was funded by research grants from the National Key R&D program of China (2017YFC1309603), National Key research and Development Program of China (2016YFA0101002), National Natural Science Foundation of China (NO. 81170736, 81570715), and Beijing science and technology project (D141107005314002). We thank all the subjects in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
All procedures performed in this study involving human participants were in accordance with the ethical standards of the Peking Union Medical College Hospital Ethics Committee.
Human and animal rights
The study was conducted in accordance with the principles of the Declaration of Helsinki of 1975, as revised in 2008.
Informed consent
Informed consent was obtained from all patients for being included in the study.
Additional information
Managed by Massimo Federici.
Rights and permissions
About this article
Cite this article
Wang, X., Wang, T., Yu, M. et al. Screening of HNF1A and HNF4A mutation and clinical phenotype analysis in a large cohort of Chinese patients with maturity-onset diabetes of the young. Acta Diabetol 56, 281–288 (2019). https://doi.org/10.1007/s00592-018-1232-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00592-018-1232-x