Abstract
The development of an efficient transformation system is required to alter the expression of symbiosis-regulated genes and to develop insertional mutagenesis in the ectomycorrhizal basidiomycete Laccaria bicolor S238N. Vegetative mycelium of this fungus was transformed by Agrobacterium tumefaciens-mediated gene transfer. The selection marker was the hygromycin resistance gene of Escherichia coli (hph) under the control of the gpd promoter from Agaricus bisporus and the CaMV 35S terminator as part of the T-DNA. PCR amplification of hph and Southern blot analyses showed that the genome of the hygromycin-resistant transformants contained the cassette. The latter proved mostly single copy and random integration of part of the transgene into the fungal genome. A. tumefaciens-mediated gene transfer should facilitate future development of insertional mutagenesis, targeted gene disruption and RNA interference technology in L. bicolor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Characterization of the primary genetic traits controlling symbiosis development and its metabolic activity, such as nutrient scavenging, transport and assimilation, will help in understanding the ecological role of ectomycorrhizal symbiosis (Martin 2001; Wiemken and Boller 2002). To understand the complex interactions that control ectomycorrhizal development, large scale EST sequencing, cDNA array analysis of gene expression and proteomics have been developed on several symbiotic associations (Laurent et al. 1999; Voiblet et al. 2001; Podila et al. 2002; Peter et al. 2003; Johansson et al. 2004; Duplessis et al. 2005). Comparisons of gene expression in free-living partners and symbiotic tissues revealed significant differences in the expression levels and have identified about 150 symbiosis-regulated fungal genes (Martin et al. 2004).
Laccaria bicolor is a common ectomycorrhizal symbiont of many temperate forest trees, among them Populus trichocarpa, the first perennial plant to have its genome sequenced. In nature and in the laboratory, L. bicolor usually grows as a heterokaryon composed of two different nuclei, each of a different mating type, but axenic haploid strains isolated from spores are available (Di Battista et al. 1996). Currently, there is sequence from approximately 2,000 ESTs and random genomic fragments deposited in GenBank (Podila et al. 2002; Peter et al. 2003). The Joint Genome Institute (JGI) is currently sequencing the genome of the ectomycorrhizal basidiomycete Laccaria bicolor S238N (Lammers et al. 2004; Martin et al. 2004).
So far the generation of null mutants for these genes has been hampered by the lack of efficient transformation systems. Screening of Laccaria transformants obtained using protoplast-PEG-based transformation (Barrett et al. 1990) and particle-bombardment-mediated method (Bills et al. 1999) showed multiple copy integrations, which is a constraint for subsequent molecular characterization of tagged genes.
A. tumefaciens-mediated transformation, besides being used for a long time to transform natural plant host (Tinland 1996), has also succeeded in transformation of human cells (Kunik et al. 2001), yeast (Bundock et al. 1995), filamentous fungi (de Groot et al. 1998) and other ectomycorrhizal basidiomycetes (Pardo et al. 2002; Hanif et al. 2002; Combier et al. 2003). Moreover, transformation with this method is mainly the result of single-copy integration events (Hellens et al. 2000).
Here we report the successful A. tumefaciens-mediated transformation of the vegetative mycelium of L. bicolor to hygromycin resistance. Southern blot analysis indicates that most transformants contain a single copy of the transgene integrated at random.
Materials and methods
Strains and plasmids
L. bicolor (Maire) Orton isolate S238N was described previously (Di Battista et al. 1996). The dikaryotic vegetative mycelium was maintained at 22°C in the dark on agar P5 medium (Martin et al. 1998). Agrobacterium tumefaciens strains LBA1100 and AGL1 were kindly supplied by Paul Hooykaas (Leiden University, Netherlands) and by Peter Romaine and Carl Schagnhaufer (Pennsylvania State University, USA), respectively. Voucher specimens are kept in the Universidad Nacional de Quilmes Culture Collection (Argentina). Plasmid pBGgHg (Chen et al. 2000) provided by Peter Romaine and Carl Schagnhaufer was used in this study because it has been proved useful in transformation of other homobasidiomycetous ectomycorrhizal fungi (Hanif et al. 2002; Combier et al. 2003). This plasmid consists of a pCAMBIA 1300 backbone with an Escherichia coli hygromycin resistance cassette (hph) driven by the Agaricus bisporus gpd promoter and terminated by the CaMV 35S terminator (Chen et al. 2000). The vector was introduced into A. tumefaciens LBA1100 and AGL-1 strains by electroporation (Pardo et al. 2002).
T-DNA transfer
A. tumefaciens-mediated transformation of L. bicolor S238N was done as described (Pardo et al. 2002). Briefly, fungal colonies were grown from several 2-mm agar plugs on cellophane membranes on P5 media at 22°C in the dark during 5 days and then transferred to P20-induction plates [P5 media with lower sugar content (glucose 2 g l−1 as the sole carbon source) supplemented by 40 mM MES, 0.5% glycerol and 200 μM acetosyringone (AS) (Aldrich, Milwaukee, WI, USA) at pH 5.3]. The colonies were then inoculated with 50 μl of an induced A. tumefaciens (pBGgHg) culture prepared according to Pardo et al. (2002). The co-cultivation plates were incubated at 22°C for 5 days in the dark; cellophane membranes containing the fungal colonies were transferred to new plates containing the selection medium [per litre: P5 media supplemented by 100 μg ml−1 cefotaxime, 100 μg ml−1 ampicillin and 100 μg ml−1 tetracycline for killing A. tumefaciens cells, and 300 μg ml−1 of HygroGold hygromycin (Invitrogen, Carlsbad, CA, USA) for selection of L. bicolor transformants] and kept at 22°C for 7–10 days in the dark.
Resistant colonies were individually harvested and isolated for vegetative propagation in P5 medium containing 300 μg ml−1 hygromycin. Four experiments, two using A. tumefaciens LBA100 and two using A. tumefaciens AGL-1 as donor strains, respectively, were done in six replicates each with 15 fungal colonies per Petri dish (9 cm diameter). Controls using non-transformed wild-type mycelium were always included.
Mycelium from the hygromycin-resistant (after three rounds of selection) and wild-type strains grown for 20 days on cellophane membranes on P5 media, with or without 150 μg ml−1 of hygromycin, respectively, was frozen in liquid nitrogen, ground to a fine powder, and genomic DNA was extracted with the DNeasy Plant Mini Kit (Qiagen, Germany). The hph resistance gene was amplified by PCR with primers HPH-F (5′-AAGCCTGAACTCACCGCGAC-3′) and HPH-R (5′-CTATTCCTTTGCCCTCGGAC-3′), which give rise to an approximately 1-kb fragment. About 50 ng of genomic DNA and about 1 ng of pBGgHg plasmid DNA, respectively, were used as template. The PCR program was as follows: initial denaturation at 94°C for 2 min, 35 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 1 min, and a final extension at 72°C for 5 min. The amplification products were purified with the Wizard PCR preps DNA purification system (Promega, Madison, WI, USA) and sequenced with the primer HPH-F, the CEQ Dye Terminator Cycle sequencing kit and the CEQ2000XL automated sequencer (Beckman Coulter, Fullerton, CA, USA). To prove the absence of contaminating A. tumefaciens cells within the resistant fungal colonies a control PCR was done with primers VIRA-F (5′-ATGAATGGAAGGTATTCACCG-3′) and VIRA-R (5′-GTTTTTGGAGCATGTCGAGTT-3′) specific for the virA gene located in the plasmid Ti. PCR conditions were as used for hph except that the annealing temperature was 55°C and 1 μl of an overnight Agrobacterium culture was used as positive control. This PCR should be negative in truly transformed fungi but positive if A. tumefaciens cells were contaminating the mycelium. Southern blot analysis was done by using an approximately 1-kb hph fragment as a probe, amplified from pBGgHg by PCR with HPH-F and HPH-R primers under the above PCR conditions. The PCR fragment was cleaned with the Wizard PCR preps DNA purification system (Promega) and labelled with alkaline phosphatase for chemiluminescent detection with the AlkPhos Direct Labelling and Detection System with CDP-Star (Amersham Biosciences, UK) according to the manufacturer's protocol. Genomic DNA (10 μg) from control and putative transformed mycelium was digested with EcoRV (Promega), which cuts once within the T-DNA but outside of the hph selection marker, and blotted onto Hybond N+ membrane (Amersham Biosciences) by alkaline transfer and baking according to the manufacturer's protocol.
Results and discussion
To test the A. tumefaciens-mediated transformation of L. bicolor, vegetative mycelium was co-cultivated with A. tumefaciens LBA1100 or AGL-1 containing pBGgHg. This construct was efficiently used for transformation of other ectomycorrhizal basidiomycetes (Hanif et al. 2002; Combier et al. 2003). In addition, the resistance level of L. bicolor to hygromycin was in a range allowing efficient selection of transformants. Co-cultivation of L. bicolor with A. tumefaciens (pBGgHg) resulted in the formation of small hygromycin-resistant colonies after 7–10 days on selective media (Fig. 1).
L. bicolor transformants (clones P1, 1, 8 and 21) and wild type (wt) grown on P5 media (a) or P5 plus 300 μg of hygromycin/ml (b) at 22°C during 7 days. Clones 1, 8 and 21 derived from co-cultivation of L. bicolor with A. tumefaciens AGL-1 and clone P1 from co-cultivation with A. tumefaciens LBA1100
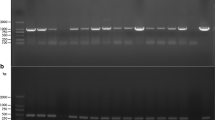
After three rounds of selection, PCR amplification using the HPH-F and HPH-R primers indicated that the hygromycin-resistant colonies contained the hph sequence (Fig. 2) (confirmed by sequencing) and thus were actually transformed. The efficiency of transformation averaged 10 and 55% when LBA1100 and AGL-1 strains were used as donors, respectively. Differences among A. tumefaciens strain efficiencies are well known and the appropriate combination of selection marker, binary vector and Agrobacterium strain is critical (Hellens et al. 2000). Transformed fungal DNA was proved to be free of A. tumefaciens DNA, as these samples were always negative in a PCR targeted to the virA gene, which is located in the plasmid Ti.
PCR analysis of L. bicolor transformants and wild type targeted to approximately 1-kb hph transgene fragment. From left to right: molecular size marker 100-bp ladder, L. bicolor wild type (wt) and transformants (P1, 1, 8 and 21), positive control with plasmid pBGgHg as template, and negative control without DNA
To determine the number of different integrations of the T-DNA, genomic DNA from transformants was digested with EcoRV and then probed with the 1-kb hph fragment from pBGgHg (Fig. 3). EcoRV cuts once within the T-DNA and the probe hybridizes to only one of the resulting fragments, thus multiple hybridizing bands would be indicative of multiple T-DNA integration events. By this criterion, transformants mostly contained single copies of T-DNA and this integration event occurred at random. The results presented in Figs. 1, 2 and 3 are representative of about 100 individual transformants from four independent experiments, taking into account the percentages of transformation obtained with each Agrobacterium strain.
Southern blot analysis of L. bicolor transformants and wild type. Total DNA (10 μg) was digested with EcoRV, blotted and probed with the approximately 1-kb hph transgene fragment. From left to right: molecular size marker λ/EcoRI/HindIII, L. bicolor wild type (wt) and transformants (clones P1, 1, 8 and 21)
The mitotic stability of part of the transferred T-DNA was tested by growing the transformants in P5 media without selection. In all cases, resistance to hygromycin was retained after this treatment.
A. tumefaciens-mediated transformation is less labor-intensive than protoplast-requiring or biolistic transformation protocols developed for Laccaria (Barrett et al. 1990; Bills et al. 1999). Moreover, single and random integration are two critical features requested for the development of large-scale insertional mutagenesis and further recovery of the T-DNA flanking sequences by PCR-based techniques (Combier et al. 2003) or plasmid rescue techniques (Leclerque et al. 2004). The transformation procedure described here is currently used to generate mycorrhiza-defective mutants of L. bicolor S238N as recently done for Hebeloma cylindrosporum (Combier et al. 2004). A precise understanding of how the symbiosis-regulated genes and proteins function and interact with each other in a cellular context requires the ability to introduce precise alterations within specific components of these genetic networks. Targeted transgenesis and RNA interference, already possible in fungal phytopathogens (Kadotani et al. 2003; Fitzgerald et al. 2004), are not yet available for the study of ectomycorrhizal symbiosis. In this respect, Agrobacterium-mediated genetic transformation of the model poplar mycorrhizal symbiont L. bicolor will probably be a useful tool to reach these goals in the future.
References
Barrett V, Dixon RK, Lemke PA (1990) Genetic transformation of a mycorrhizal fungus. Appl Microbiol Biotechnol 33:313–316
Bills S, Podila G, Hiremath S (1999) Genetic engineering of a fungus Laccaria bicolor for use as a biological control agent. Mycologia 9:237–242
Bundock P, Dulk-Ras A, Beijersbergen A, Hooykaas PJJ (1995) Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J 14:3206–3214
Chen X, Stone M, Schlagnhaufer C, Romaine CP (2000) A fruiting body tissue method for efficient Agrobacterium-mediated transformation of Agaricus bisporus. Appl Environ Microbiol 66:4510–4513
Combier JP, Melayah D, Raffier C, Gay G, Marmeisse R (2003) Agrobacterium tumefaciens-mediated transformation as a tool for insertional mutagenesis in the symbiotic ectomycorrhizal fungus Hebeloma cylindrosporum. FEMS Microbiol Lett 220:141–148
Combier JP, Melayah D, Raffier C, Pépin R, Marmeisse R, Gay G (2004) Nonmycorrhizal (Myc−) mutants of Hebeloma cylindrosporum obtained through insertional mutagenesis. Mol Plant-Microb Interact 17:1029–1038
de Groot MJA, Bundock P, Hooykaas PJJ, Beirjesbrgen AGM (1998) Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16:839–842
Di Battista C, Selosse MA, Bouchard D, Stenstrøm E, Le Tacon F (1996) Variation in symbiotic efficiency, phenotypic characters and ploidy level among different isolates of the ectomycorrhizal basidiomycete Laccaria bicolor strain S238. Mycol Res 100:1315–1324
Duplessis S, Courty P, Tagu D, Martin F (2005) Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus. New Phytol 165:599–611
Fitzgerald A, van Kan J, Plummer K (2004) Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet Biol 41:963–971
Hanif M, Pardo AG, Gorfer M, Raudaskoski M (2002) T-DNA transfer and integration in the ectomycorrhizal fungus Suillus bovinus using hygromycin B as a selectable marker. Curr Genet 41:183–188
Hellens R, Mullineaux P, Klee H (2000) Technical focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5:446–451
Johansson T, Le Quéré A, Ahren D, Søderstrøm B, Erlandsson R, Lundeberg J, Uhlén M, Tunlid A (2004) Transcriptional responses of Paxillus involutus and Betula pendula during formation of ectomycorrhizal root tissue. Mol Plant-Microb Interact 17:202–215
Kadotani N, Nakayashiki H, Tosa Y, Mayama S (2003) RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol Plant-Microb Interact 16:769–776
Kunik T, Tzfira T, Kapulnik Y, Gafni Y, Dingwall C, Citovsky V (2001) Genetic transformation of HeLa cells by Agrobacterium. Proc Natl Acad Sci U S A 98:1871–1876
Lammers P, Tuskan G, DiFazio S, Podila G, Martin F (2004) Mycorrhizal symbionts of Populus to be sequenced by the United States Department of Energy's Joint Genome Institute. Mycorrhiza 14:63–64
Laurent P, Voilet C, Tagu D, de Carvalho D, Nehls U, De Bellis R, Ballestrini R, Bauw G, Bonfante P, Martin F (1999) A novel class of ectomycorrhiza-regulated cell wall polypeptides in Pisolithus tinctorius. Mol Plant-Microb Interact 12:862–871
Leclerque A, Wan H, Abschütz A, Chen S, Mitina G, Zimmermann H, Schairer H (2004) Agrobacterium-mediated insertional mutagenesis (AIM) of the entomophatogenic fungus Beauveria bassiana. Curr Genet 45:111–119
Martin F (2001) Frontiers in molecular mycorrhizal research—genes, loci, dots and spins. New Phytol 150:499–505
Martin F, Delaruelle C, Ivory M (1998) Genetic variability in intergenic spacers of ribosomal DNA in Pisolithus isolates associated with pine, Eucalyptus and Afzelia in lowland Kenian forest. New Phytol 139:341–352
Martin F, Tuskan GA, DiFazio SP, Lammers P, Newcombe G, Podila GK (2004) Symbiotic sequencing for the Populus mesocosm. New Phytol 161:330–335
Pardo AG, Hanif M, Raudaskoski M, Gorfer M (2002) Genetic transformation of ectomycorrhizal fungi mediated by Agrobacterium tumefaciens. Mycol Res 106:132–137
Peter M, Courty PE, Kohler A, Delaruelle C, Martin D, Tagu D, Frey-Klett P, Duplessis S, Chalot M, Podila G, Martin F (2003) Analysis of expressed sequence tags from the ectomycorrhizal basidiomycetes Laccaria bicolor and Pisolithus microcarpus. New Phytol 159:117–129
Podila GK, Zheng J, Balasubramanian S, Sundaram S, Hieremath S, Brand J, Hymes M (2002) Molecular interactions in ectomycorrhizas: identification of fungal genes involved in early symbiotic interactions between Laccaria bicolor and red pine. Plant Soil 244:117–128
Tinland B (1996) The integration of T-DNA in plant genomes. Trends Plant Sci 1:178–184
Voiblet C, Duplessis S, Encelot N, Martin F (2001) Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. Plant J 25:181–191
Wiemken V, Boller T (2002) Ectomycorrhiza: gene expression, metabolism and the wood-wide web. Curr Opin Plant Biol 5:355–361
Acknowledgements
We are grateful to Peter Romaine and Carl Schlagnhaufer for providing A. tumefaciens strain AGL-1 and plasmid pBGgHg and to Paul Hooykaas for providing A. tumefaciens strain LBA1100. A.G.P. is a member of the Scientific Research Career of CONICET (Argentina). This programme was financed by INRA, Région de Lorraine, PICT-ANPCyT, UNQ and the France–Argentina ECOS-Sud grants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kemppainen, M., Circosta, A., Tagu, D. et al. Agrobacterium-mediated transformation of the ectomycorrhizal symbiont Laccaria bicolor S238N. Mycorrhiza 16, 19–22 (2005). https://doi.org/10.1007/s00572-005-0008-7
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-005-0008-7