Abstract
Correct segmentation of stroke lesions from magnetic resonance imaging (MRI) is crucial for neurologists and patients. However, manual segmentation relies on expert experience and is time-consuming. The complicated stroke evolution phase and the limited samples pose challenges for automatic segmentation. In this study, we propose a novel deep convolutional neural network (Res-CNN) to automatically segment acute ischemic stroke lesions from multi-modality MRIs. Our network draws on U-shape structure, and we embed residual unit into network. In Res-CNN, we use residual unit to alleviate the degradation problem and use multi-modality to exploit the complementary information in MRIs. Before training the model, we use data fusion and data augmentation methods to increase the number of training images. Seven neural networks are extensively evaluated on two acute ischemic stroke datasets. Res-CNN shows good performance compared with other six networks both in single modality and multi-modality. Furthermore, compared with the gold standard segmentation manually labeled by two neurologists on a local test dataset, our network achieves the best results in seven neural networks. The average Dice coefficient and Hausdorff distance of our method are 74.20% and 2.33 mm, respectively. Our proposed network may provide a useful tool for segmentation lesion of acute ischemic stroke.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The World Health Organization (WHO) is an assessment of the global disease status which shows that stroke is the second leading cause of death all over the world. Stroke prevents blood to reach brain regions; it is a cause of death and disability. Accurate quantification and automatic segmentation of the stroke lesions can provide insightful information for neurologists. It is an important metrics for planning treatment strategies, monitoring disease progression and predicting patient outcome [1]. However, stroke is a complex small blood vessel disease. Stroke not only has a variety of types (ischemic stroke, sub-acute ischemic stroke, ischemic penumbra, chronic stroke), but also has many similar diseases, such as white matter hyperintensities (WMHs), multiple sclerosis (MS) and so on [2,3,4,5,6]. In addition, the location, shape and size of stroke lesions are variable. All of these pose challenges for stroke lesions segmentation.
In the last decade, manual segmentation of stroke lesions plays an important role in assessing clinical prognosis. It can help track down disease evolution phase and assess the treatment effectiveness [1]. However, manual segmentation is a tedious process. For neuroscientists, it is time-consuming and laborious to segment large numbers of MRI images. The quality of segmentation depends on the subjective experience of neuroscientists [7]. In this regard, automatic segmentation method shows great advantages. It can provide a series of consistent measurements and quantitative analyses for image segmentation. For example, Cardoso et al. [8] proposed a template-based multimodal and applied it to brain image classification. They offered a robust way to identify abnormal patterns in medical images. Ledig et al. [9] proposed a framework for segmentation of magnetic resonance brain images. They used multi-atlas to improve the performance of the framework. Rekik et al. [10] proposed a common categorization pattern to analyze the ischemic stroke images, which was beyond those basic thresholding techniques. Hevia et al. [11] combined the region segmentation and edge detection method as a segmentation technology, which can segment cerebral infarction regions from diffusion-weighted imaging (DWI) images in acute ischemic stroke. Most of these methods are unsupervised methods or classification methods, which depend on multi-atlas labels or artificial corrections.
In recent years, many supervised and efficient segmentation methods are proposed, which are based on machine learning methods and deep learning methods [12]. These methods are superior to the conventional methods in the tasks of segmentation. For example, Rajini et al. presented a method to detection of ischemic stroke from computed tomography (CT) images. They used support vector machine (SVM), k-nearest neighbor (k-NN), artificial neural network (ANN) to classify normal brain and ischemic brain, respectively. The method was demonstrated to improve efficiency and accuracy of clinical practice [13]. Griffis et al. [14] used probabilistic tissue segmentation and image algebra to create feature maps encoding information about missing and abnormal tissue in T1-weighted (T1-w) images and used naive Bayes classifier to identify stroke lesions. Chen et al. proposed a semantic segmentation network (EDD Net), which was based on convolutional neural network (CNN). This network was used to segment stroke lesions in DWI images and tried to achieve optimal lesion segmentation in all scales [15]. Zhang et al. proposed a fully convolutional neural network (FCN) to segment stroke lesions from DWI images. The network could utilize contextual information and learn discriminative features in an end-to-end way [16]. Liu et al. proposed a residual-structured fully convolutional network (Res-FCN) to automatically segment ischemic stroke lesions from multi-modality MRIs. In Res-FCN, the residual block could capture features from large receptive fields for the network [17].
Different input modality sequences are used in lesions segmentation methods. The selections of modality sequences are directly related to the performance of segmentation method. The common MRI sequences include T1, T1-w, T2, T2-weighted (T2-w), DWI, apparent diffusion coefficient (ADC) and fluid attenuation inversion recovery (FLAIR). Chen et al. only used DWI sequence as input to segment acute ischemic lesions. Their network was validated on a large real clinical dataset. The average Dice coefficient (DC) of segmentation results was 0.67 [15]. Havaei et al. proposed a CNN to segment sub-acute and ischemic lesions. Their network used DWI, FLAIR and T2 as inputs, and the DC of two segmentation tasks was 0.35 and 0.54, respectively [18]. Kamnitsas et al. and Liu et al. used two or more than two MRI sequences as inputs for stroke segmentation lesions [17, 19]. In their experiments, these modality sequences were more conducive to the improvement of network performance than other sequences.
There is a serious change about lesion density, size and shape during the first hours or days of stroke, which determine the state of lesions in MRI sequences [20, 21]. Most stroke segmentation methods pay more attention to change network structure. However, most of these methods rarely analyze the characteristics of MRI sequences, which is an important factor in stroke evolution phase. Only a few stroke studies have given the details of evolution phases as inclusion and exclusion criteria [22, 23]. In addition, the traditional CNN models usually need thousands of training images, which is beyond the scope of medical image analysis tasks. As it is known, for small sample datasets, it can cause the vanishing gradient problem when a network has deep layers.
The goal of this study is to validate a robust and automatic CNN framework for acute ischemic stroke lesion segmentation, which is based on multi-modality MRI sequences. To achieve this goal, we investigate the performance of our method across two different datasets. In this paper, our main contributions can be summarized as follows:
-
(1)
Our method, called Res-CNN, combines similar U-shape architecture with residual units. This network could alleviate the vanishing gradient problem.
-
(2)
According to the characteristics of MRI modalities in acute ischemic stroke evolution phase, we fuse T2 to DWI (DWI-T2) as a complementary modality and we use augmentation methods to increase the number of training images. We use DWI and DWI-T2 as input multi-modality to improve the performance of lesion segmentation.
-
(3)
Extensive experiments on two acute ischemic stroke datasets: a public dataset stroke penumbra estimation (SPES) of Ischemic Stroke Lesion Segmentation (ISLES) 2015 challenge and a local hospital clinical dataset (LHC). Compared with other methods, our network achieves the state-of-the-art performance on SPES challenge, and it is closed to the segmentations from neurologists in LHC.
The rest of this paper is organized as follows. We first describe two MRI datasets, describe data preprocessing and augmentation methods, and then, we detail the proposed Res-CNN network framework in Sect. 2. Experiments and results are demonstrated in Sect. 3. We further discuss and analyze our study in Sect. 4. Finally, conclusions are drawn in Sect. 5.
2 Material and methods
2.1 Datasets
Our goal is to segment the lesions of acute ischemic stroke with conventional image sequences. In order to validate the robustness and generalizability of the method, the proposed Res-CNN framework is only trained on SPES dataset and tested on SPES and LHC datasets, respectively. All patients in both two datasets were treated for acute ischemic stroke.
For SPES, stroke MRIs are scanned on either 3.0T or 1.5T MRI system (Siemens Magnetom Trio or Siemens Magnetom Avanto) [24]. Each patient has seven medical modality images [T1 contrast-enhanced (T1c), T2, DWI, cerebral blood flow (CBF), cerebral blood volume (CBV), time-to-peak (TTP) and time-to-max (Tmax)]. In SPES, all modality sequences of a patient correspond to the infarct core label. The labels are the ground truth segmentation map which are manually drawn on DWI images.
In SPES, coregistered and preprocessed methods were preprocessed by the organizers [24], including intensity range standardization, skull-stripped sequences, constant resolution, bias field correction, isotropic voxel-spacing and affine registration to montreal neurological institute (MNI) space [25]. There is no further processing that is applied to the MRIs of SPES. In our study, we used 30 brain samples with ground truth infarct core labels in SPES dataset and with DWI and T2 conventional modalities.
The MRIs in LHC were a subset of acute ischemic stroke patients from a local hospital stroke in 2016. The scans were obtained from Philips Achieve 3.0T MRI system with following acquisition parameters: field strength: 3.0T; matrix size: 230 × 230 × 18; slice thickness: 6 mm; field of view: 230 mm × 230 mm; slices: 18; slice spacing: 1.0–1.5 mm; echo time: 87 ms; pixel size in x–y plane: 0.9 × 0.9 or 1.51 × 1.90 mm; repetition time: 23 ms. All samples in LHC are used as test data with DWI and T2 modalities. The brain of infants and adolescents is still growing, and the brain structure is not stable. There are some similar brain diseases for the elderly, such as WMH, MS, and other small vessel diseases [6] are commonly observed in MRIs [26]. Distinguishing similar diseases is not the purpose of our study. In LHC, we excluded patients at younger or older age, and the age of our patients is between 20 and 50. Finally, we got 29 samples. Each sample contained 18 2D image slices. The stroke infarcts were found in 3–4 slices in each sample. We selected 3 slices from each sample, which contained stroke infarcts. In the end, we got 87 images as testing images.
2.2 Data preprocessing
The MRIs in LHC were acquired from different scanners under different parameters settings in routine clinical examination. Images were stored in DICOM format with DWI, T1 and T2 modality sequences. Most raw MRIs contain noises, which makes rendering appear dusty or hazy. Several pre-processing steps must be conducted before experiments. First, we used SPM12 software to convert DICOM format images into NIfTI format images. Second, during the early onset of acute ischemic stroke, DWI modality is a sensitive and specific modality for diagnosis [27,28,29,30]. Images with T2 and T1 modalities were coregistered to the DWI images by using a six-parameter rigid body registration method [31]. Third, MRI sequences were skull-stripped by MRIcron software (BET2) [32]. We extracted brain with manual correction, and then, we re-coregistered MRI sequences to DWI again. In the end, all MRIs were smoothed by Gaussian method.
2.3 Multi-modality and data augmentation
In the field of MRIs analysis, multiple MRIs are used to examine different lesion tissues. The information about multi-modality images is complementary, and it can provide more comprehensive information for diagnosis, such as Moeskops et al. [33] used T1, T1-IR and T2-FLAIR imaging modalities in automatic brain structure segmentation, Menze et al. 2015 used T1, T1c, T2 and T2-FLAIR imaging modalities in brain tumor segmentation task [24], Maier et al. [23] used T1-w, T2-w, DWI and FLAIR imaging modalities in brain lesion analysis. The performances of these methods are much better than that of single-modality methods in the image analysis tasks.
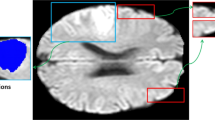
In the process of clinical diagnosis, some MRI modalities play major roles in diagnosis of the acute ischemic stroke [27, 30, 34]. In the early stages of stroke, DWI modality is more sensitive in the diagnosis of stroke than other MRI sequences [27, 30]. T2 modality usually is used to distinguish the ischemic core and other impaired tissues [30, 34]. We investigate DWI and T2 modalities, and we fuse T2 image to DWI image space to generate a complementary image using structured and sparse canonical correlation analysis (ssCCA) technique [35], the new image is DWI-T2. From Fig. 1, it can be seen that the lesion tissues are enhanced in DWI-T2 modality.
In most MRI analysis tasks, compared with other vision datasets, the sample sizes of MRI datasets are smaller, such as a large-scale ontology of images dataset “ImageNet” [36] and the dataset in skin lesion classification [37]. Most medical MRI datasets have a limited number of samples, and the number of scanned lesions is also limited. However, deep learning models need a large number of images for training a huge number of parameters in the model. If training data are generated by slicing images or lesion instance patches, the amount of samples is far from the requirements for deep learning model. Chatfield et al. showed that data augmentation techniques can be applied to deep learning methods, and those techniques result in an analogous performance boost [38, 39]. In our study, all samples are in 3D form. Each 3D sample has more than 60 2D image slices in SPES. However, not all image slices contain stroke infarcts. We excluded the slices of hemorrhagic stroke. In addition, we also excluded the slices, which do not contain stroke infarct in MRI sequence. Of 30 samples, one only contains 14 slices of stroke infarct. For keeping the balance on the number of image slices for each sample, we selected 14 2D images from each sample, which contained stroke infarcts. In the end, we got 420 (14 × 30) 2D image slices. In our study, for increasing the number of training images, we performed data augmentation on training 2D images. Each image was rotated 0°, 90°, 180° and 270°, respectively. Then, we used flip and mirror mapping methods to augment the number of images [40, 41]. One image becomes eight images. However, there are redundant images in the flipped images. For example, an image was rotated 180°, and then, it turned into the original image after mirror mapping operation. We excluded redundant images. As a result, we can obtain 1680 2D images in each modality. It should note that data augmentation only was performed on training images in SPES. The number of test images did not augment. For our experiments, we randomly select 25 samples as cross-validation training samples and the remaining five as testing samples in SPES. So in each experiment, there are about 1344 2D image slices for training and 90 2D image slices for testing.
2.4 Res-CNN network
As reported in the study of He and Sun, when deep networks start converging, the degradation problem has been exposed [42, 43]. In this study, we proposed a novel automatic segmentation network (Res-CNN) to alleviate the degradation problem. The architecture of Res-CNN is illustrated in Fig. 2. The detail of network structure is shown in Table 1. It composes of 10 convolution layers, 4 residual units, 4 concatenation layers (Concat), 4 deconvolution layers (Deconv), and some batch normalization layers (BN), leaky rectified linear units (LReLU) [44]. Some BN and LReLU are embedded in the residual units. BN layers are used to reduce internal covariance shift, which would help improve the performance of the training process. LReLU layers are utilized as the activation functions for nonlinear transformation. LReLU layers allow a small, nonzero gradient when the unit is not active, i.e., \(f(x)= \left\{ \begin{array}{lll} x, \ \ {\text{if}} \ \ x>0 \\ 0.01x, \ \ {\text{otherwise}} \end{array}. \right.\)
Res-CNN consists of two symmetrical paths. The left side is a contracting path, which is used to obtain context information. The right side is an expansive path, which is used to achieve precise positioning. The architecture of contracting path mainly follows that of the typical convolutional network. The lowest layer of contracting path obtains the resolution size of input convolution size by a factor of 1/8. The expansive path of Res-CNN not only extracts features, but also expands the spatial support of lower resolution features by up-sampling. The forward features are copied from the contracting part of the network to expansive part by horizontal connections [45]. We use horizontal connections to gather fine-grained details, otherwise it would be lost in the contracting path. This method also improves the quality of final contour prediction. In our network, every layer in the expansive path consists of an up-sampling of the feature map. A deconvolution layer follows an up-sampling with a 2 × 2 kernel size. The concatenation layers have the correspondingly feature map from contracting path to expansive path. Finally, the feature maps were computed by three convolution layers. The kernel size of the first convolution layers was set to 3 × 3; the kernel size of the last convolution layer was set to 1 × 1. We obtain the probability of a pixel belonging to the foreground region or background region with a wise sigmoid function and convert the resulting output map. We produce the segmentation result with the same size as the input MRI.
The architecture of residual unit is shown in Fig. 3, and there are two pathways for information propagating. One is an direct pathway from \(R_l\) to \(R_{l+1}\), and another is an indirect pathway with successive layers from \(R_l\) to \(R_{l+1}\). Two pathways are merged before output. In each residual unit, there has an input feature (\(R_l\)) and a transformed feature (includes two repeated BNs, LReLUs and convolution layers). Then, two features are integrated together as the feature \(R_{l+1}\), and information can be transmitted directly in forward and backward directions [46]. The residual unit in Fig. 3 can be expressed in the following general form:
where \(R_l\) is the input feature and \(R_{l+1}\) is the output result of the l-th residual unit. \(W_l=\{W_{l,k}|1\le k\le K\}\) is a set of weights, which is associated with the l-th residual unit. The F() is the residual function.
With Eq. (1), the \(R_L\) (\(L>l \ge 1\)) can be derived in a recursive way as follows:
where l is a shallower unit and L is a deeper unit. \(\sum _{i=l}^{L-1}F()\) denotes the summarized units between l and \(L-1\), it is a residual function. Equation (2) shows that any deeper feature \(R_L\) can be expressed as any superimposed \(R_l\) with a residual function \(\sum _{i=l}^{L-1}F()\). According to the chain rule of back propagation [47], we get the derivatives as:
where \(\varepsilon\) denotes the loss function of residual units. \(\frac{\partial \varepsilon }{\partial R_l}\) represents information without concerning any weight layers, it ensures that information is directly propagated back to any shallower unit l. \(\frac{\partial }{\partial R_l}\sum _{i=l}^{L-1}F(R_i,W_i)\) represents that propagation passes through the weight layers. Equations (2) and (3) reveal that the information can be directly propagated backward and forward from one unit to another in the network [46].
In this study, we used DWI and DWI-T2 as input, and the output segment images were activated by the sigmoid function, which outputs the probability of each pixel belonging to the foreground or the background. However, compared with the background, lesions occur in very small region of MRIs. This can lead to a serious bias in the prediction results [48]. In this study, we only care about the lesions of segmentation results, which are not background. We use S(X, Y) to measure the parts of lesions, which overlap between the predicted segmentation and the reference ground truth. S(X, Y) can be written as:
where X and Y denote the ground truth image and predicted segmentation image, respectively. For each element \(X_{ij}\) in X, if it belongs to the area of lesion, \(X_{ij}\) is set to 1, otherwise \(X_{ij}\) is set to 0. For each element \(Y_{ij}\) in Y, if it belongs to the area of predicted lesion, \(Y_{ij}\) is set to 1, otherwise \(Y_{ij}\) is set to 0. Z also denotes an image, in which the value of the element in position (i, j) will be set to 1 only when both \(X_{ij}\) and \(Y_{ij}\) are equal to 1. \(Z_{ij}\) is an element in Z. That is to say \(Z_{ij}= \lfloor \frac{X_{ij}+Y_{ij}}{2}\rfloor\). \(\left| X \right|\) is a function which counts the number of elements with ’1’ in image X.
If there is no lesion in X and Y, both \(\left| X \right|\) and \(\left| Y \right|\) are 0. In this case, the denominator in Eq. (4) will be 0. To solve the problem, we add a small k in denominator and numerator of Eq. (4). Generally, both \(\left| X \right|\) and \(\left| Y \right|\) are more than 100 if they are not equal to 0. So we set the value of k in Eq. (4) to 0.5, which has very little influence on the result of S(X, Y).
In this study, we proposed a novel loss function L(TX, PY) which can be written as:
where N is the number of 2D images. TX is a set of ground truth images, and PY is a set of predicted segmentation images. \({\text{TX}}_i\) and \({\text{PY}}_i\) are images in TX and PY, respectively. \({\text{TX}}_i\) is the ground truth of the ith image, \({\text{PY}}_i\) is the predicted result of the ith image.
2.5 Implementation details
In this study, we proposed a Res-CNN framework to segment acute ischemic stroke lesions. In order to utilize more contextual information, we used DWI and DWI-T2 modality sequences to strengthen the pixel feature information. All MRIs are unified with the 160 × 160 pixel size using the ITK tools.Footnote 1 The multi-modality architecture used in our neural network is shown in Fig. 4. The network uses DWI and DWI-T2 modality sequences as inputs.
Our network was implemented using Python based on Keras library. We chose six other medical image segmentation models for comparison, including the U-net [45], EDD Net [15], FCN [49], FC_ResNet (Fully Convolutional Residual Network) [50], Res-FCN [17] and FCN + FCN_ResNet [51]. Since each network has its own characteristics, it is difficult to completely reproduce the original model for different databases. When adapting the candidate network into our dataset, we used the same input image size. All comparison methods retain their original network structure. More specifically, the adapted U-net has concatenations between left-side layers and right-side layers. The adapted EDD Net retains the featured deconvolution and unpooling layers. The adapted FCN network is still in the fully convolutional configurations. The adapted FC_ResNets preserves residual blocks and additional shortcut paths. The adapted Res-FCN preserves the unique structures (Global Convolutional Network and Boundary Refinement) based on FCN. The adapted FCN + FCN_ResNet uses a FCN to obtain pre-normalized images and uses FCN_ResNet to generate a segmentation prediction. No post-processing operations are used in any architecture.
3 Experiments and results
3.1 Evaluation metrics
In this study, we used two metrics to evaluate our method on two datasets: Dice coefficient (DC) [52] and Hausdorff distance (HD) [53, 54].
DC is main metric in the segmentation task. It is a statistic used for computing the similarity of two sets. DC score denotes the spatial overlap between the segmentation with the reference ground truth. A larger value indicates superior segmentation accuracy. The DC value is given as:
where A is the set of reference ground truths and B is the set of segmentation results. HD is another metric, which measures the maximum surface distance between two sets (the prediction segmentation results and the ground truth). It is defined as follows:
where \(A_{\text{s}}\) and \(B_{\text{s}}\) are two nonempty sets of surface points, and a and b are the points of \(A_{\text{s}}\) and \(B_{\text{s}}\) sets, respectively. d(a, b) or d(b, a) indicates the Euclidean distance between the points a and b. s is the number of points in two sets A and B (\(s\ge 1\)). The lower HD value implies the better segmentation performance.
3.2 Multi-modality and segmentation methods
To evaluate the efficacy of multi-modality, we test the signal-modality and multi-modality on Res-CNN and other six networks, respectively: U-net, EDD Net, FCN, FC_ResNet, Res-FCN and FCN + FCN_ResNet. We performed the experiments five times. At each time, we randomly selected 25 samples as cross-validation training samples and the remaining five as testing samples in SPES. The training samples were transformed to 2D image slices. Then, the 2D training images were screened, and we performed data augmentation on 2D training images. The method of data augmentation is explained in Sect. 2.3. In each experiment, there are about 1344 2D image slices for training and 90 2D image slices for testing. All experiments were conducted under fair conditions, including the same data augmentation strategies. All experiments used the same images for training and testing, respectively. The results of the segmentation are shown in Table 2. In general, the DC scores and HD values of multi-modality are better than those of the single modality of seven methods. Compared with the other six methods, in terms of the main metric DC scores our network has the best performance both in single modality and multi-modality. For single modality, the performance of EDD Net is ranked the second. The difference of HD values between EDD Net and our network is very small. However, the DC score of our network is almost 3% higher than that of EDD Net. For multi-modality, the performance of FCN + FCN_ResNet is ranked the second. The HD value of FCN + FCN_ResNet is lower than that of ours. However, the main metric DC score of our model is 4% higher than that of FCN + FCN_ResNet, which means that our network has a better segmentation performance.
3.3 Comparison with manual segmentation
In order to investigate the relative accuracy of our proposed segmentation network, we used the best weight of each model (U-net, EDD Net, FCN, FC_ResNet, Res-FCN, FCN + FCN_ResNet and our network) to test LHC dataset, respectively. Then, we compared the segmentation results with gold standard which were manually labeled by two experienced neurologists on DWI modality. In the testing of LHC dataset, we used the 2D MRI images as testing inputs, and used DC and HD as metrics.
We compared the lesion segment images generated by each method with two manual references, respectively. The test results are shown in Table 3. Obviously, the results of Res-CNN achieved better performance than that of other models. As the main evaluation metric of image segmentation, the DC scores of the six methods on multi-modality are better than those of single modality, except the method FCN. In both single modality and multi-modality, the DC scores of our network achieve the highest scores, which indicates our network has better segmentation performance than other six methods. The HD value of our network is 0.01 mm higher than that of FCN + FCN_ResNet in single modality. However, our network gets the highest DC values among all 7 methods both in single modality and multi-modality. In conclusion, the segmentation results of our network have higher agreement with the two manual labels.
Figure 5 shows the segmentation results of 6 out of 54 test images, which are the results of segmentation methods and manual references. Compared with manual references, most segmentation methods can segment large lesions well. From top to bottom, the first row is original images. The rows 2–7 are restored images which are segmented by 7 methods. Last 2 rows are gold standard manual labels which are segmented by two neurologists. The red arrows point to true lesions in the original images and the blue arrows point to noisy points. It can be observed from the contrast images that most methods can well identify the location of lesions. However, there is a difference in the number and size of the lesions for 7 methods. As shown in the second row, the segmentation result of U-net has lots of noisy points. FCN has a good segmentation result for large lesions as shown column 1–5, but not good on small or multiple lesions as shown in column 6. FC_ResNet is an extension of FCN and residual units. The segmentation lesions of FC_ResNet are much larger than FCN, and FC_ResNet is more sensitive to small lesions than FCN, which is shown in the last column of the 4th and fifth rows. It can be seen from Table 2 and Table 3 that the performance of EDD Net, Res-FCN and FCN + FCN_ResNet is close to that of our network. However, in these 6 images, Res-FCN and FCN + FCN_ResNet show more noisy points than EDD Net and our network. As shown in the last column of Fig. 5, for multiple lesions, EDD Net, Res-FCN and FCN + FCN_ResNet cannot detect all lesions. Compared with EDD Net, Res-FCN and FCN + FCN_ResNet , our network performs better in segmentation of mutiple lesions. As shown in the last 3 rows of Fig. 5, the segmentation results of the our network are the closest to the manual references.
3.4 The robustness of the networks
As shown in Table 3, we get the segmentation performance of seven networks on multi-modality MRIs of LHC. In descriptive statistics, we use a box plot (boxplot) to graphically depict the test results. The spacing between different parts of the box indicates the degree of dispersion in the data, and the individual points are shown as outliers. With in the box, bold line denotes the median value, the first quartile is represented by the lower line and the third quartile is represented by the upper line [55]. Figure 6 shows the distribution of DC values across 0 and 1, and Fig. 7 shows the distribution of HD values between 0 and 4. As shown in Fig. 6, compared with the other 6 methods, the median line of the box of our network is higher and the length of the whisker plots box is shorter, which means that Res-CNN has more accurate segmentation results on the same test set. The height of our network box is much shorter than other six boxes, which indicates that the dispersion of segmentation results is more concentrated. As shown in Fig. 7, the box height of FCN, FCN_ResNet, Res-FCN, FCN + FCN_ResNet and our network is similar. However, the median line and lower line of our network’s box are lower than those of other four boxes, which implies that the segmentation results of our network are more proximity with the labels provided by neurologists.
4 Discussion
In this paper, we have proposed a Res-CNN network to segment the acute ischemic stroke lesions using multi-modality MRI sequences. It achieves better performance compared to the six classical CNN methods. We have done extensive experiments to verify the effect of the multi-modality in our network.
The architecture of Res-CNN mainly follows the U-shape which is based on CNN network proposed by Ronneberger et al. in 2015. In U-shape architecture, the authors used up-sampling operators to replace unpooling operators and used skip connections to directly connect opposing contracts. The loss rate of the model can be greatly reduced and thus the performance increases effectively. In our network, we used residual units replace the down-sampling in contracting path. The residual unit is used to optimize the architecture of each convolution layer. It not only makes network easer gradient backpropagation, but also improves optimization convergence speed. Rectified linear unit (ReLU) [44] is an important part of original U-net. However, ReLU units can be fragile during training. When a large gradient flows through, ReLU may cause neurons never active, which means that ReLU units can irreversibly die during training. In our network, we used leaky rectified linear unit (LReLU) instead of ReLU. LReLU allows a small, nonzero gradient. When a unit is not active, LReLU sacrifices hard-zero sparsity for a gradient which is potentially more robust during optimization [56, 57]. At the same time, we used concatenation to reduce the complexity of the network architecture, it does not have negative impact on the performance of the network.
In clinical practice, it is very important for neurologists and patients to detect the location, shape and size of the lesions. In the fusion terminology, a “modality” is defined as a single image contrast [35]. Different modality sequences play different roles in the diagnosis process. In our study, there are three single-modalities: DWI, T2 and DWI − T2. DWI + T2 denotes a multi-modality combining DWI and T2, and DWI + (DWI − T2) denotes a multi-modality combining DWI and DWI-T2. In our experiment, multi-modality sequences DWI + (DWI − T2) are selected as inputs in our network Res-CNN. As shown in Tables 2 and 3, in contrast to single modality, multi-modality helps to improve the performance of the segmentation method in most time. In our experiments, the advantage of using two image modalities can reduce the effects of distortions and noises which are found in a single modality. As shown in Figs. 8 and 9, we compared the DC scores of single modality and multi-modality in two datasets based on Res-CNN. On the whole, the height of red box of multi-modality is little shorter than that of blue box of single modality. The length of the whisker plots box of multi-modality is much shorter, and the median values in the red box is higher. We also compared the performance of different modalities in our network, which is shown in Table 4. In our experiments, compared with DWI, DWI − T2 and DWI + T2, we choose multi-modality which has a better performance both in two datasets.
There are some small lesions in LHC, as shown in Fig. 5. Compared with other methods, EDD Net and our network have a better performance on small lesion segmentation. However, there is still a far distance from perfect segmentation. In image segmentation tasks, U-net, FCN, FC_ResNet, Res-FCN and FCN + FCN_ResNet use the bilinear interpolation strategy as the sampled method to implement coarse feature mapping. However, the bilinear interpolation strategy makes network difficult to reconstruct small lesions based on weak activation [15]. To reduce the impact of small lesions, EDD Net takes the recorded pooling masks and the unpooling strategy. In Res-CNN, we used the residual unit and concatenation strategy to handle this problem. In the future work, we should pay attention to the small lesion segmentation. We would draw unsupervised spectral feature selection (USFS) method to extract more local features from MRIs [58].
In the experimental stage, we have recorded the computational cost of each model. All models were trained and tested on NVIDIA GeForce Titan X Pascal CUDA GPU processor. The results are shown in Table 5. Although the average training times of different models are different, the time gaps between different models on testing are very small.
5 Conclusions
In this paper, we proposed a Res-CNN network to automatically segment acute stroke lesions in multi-modality MRIs. We analyzed the characteristics of the MRI modality based on ischemic stroke evolution phase, and we used multi-modality elegantly integrated into our network to improve the model capability. We evaluated the performance of the network on both datasets. The experimental results showed that our method outperformed other well established and state-of-the-art segment methods in terms of overlap with the ground truth on both datasets. In the future, further improvements could be made by collecting more MRI samples and using 3D convolutions. We would also try to analyze stroke disease by combining brain networks and hyper-graph techniques [59,60,61,62].
Notes
References
Polman CH, Reingold SC, Edan G, Filippi M, Hartung H-P, Kappos L, Lublin FD, Metz LM, McFarland HF, O’Connor PW (2005) Diagnostic criteria for multiple sclerosis: 2005 revisions to the “mcdonald criteria”. Ann Neurol 58(6):840–846
Chyzhyk D (2015) An active learning approach for stroke lesion segmentation on multimodal mri data. Neurocomputing 150:26–36
Jacobsen C, Hagemeier J, Myhr KM, Nyland H, Lode K, Bergsland N, Ramasamy DP, Dalaker TO, Larsen JP, Farbu E (2014) Brain atrophy and disability progression in multiple sclerosis patients: a 10-year follow-up study. J Neurol Neurosurg Psychiatr 85(10):1109
McKinley R, Häni L, Wiest R, Reyes M (2015) Segmenting the ischemic penumbra: a decision forest approach with automatic threshold finding. In: International workshop on Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries, pp 275–83
Neumann AB, Jonsdottir KY, Mouridsen K, Hjort N, Gyldensted C, Bizzi A, Fiehler J, Gasparotti R, Gillard JH, Hermier M (2009) Interrater agreement for final infarct mri lesion delineation. Stroke 40(12):3768–3771
Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, Lindley RI, O’Brien JT, Barkhof F, Benavente OR (2013) Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 12(8):822–838
Grimaud J, Lai M, Thorpe J, Adeleine P, Wang L, Barker GJ, Plummer DL, Tofts PS, Mcdonald WI, Miller DH (1996) Quantification of mri lesion load in multiple sclerosis: a comparison of three computer-assisted techniques. Magn Reson Imaging 14(5):495–505
Cardoso MJ, Sudre CH, Modat M, Ourselin S (2015) Template-based multimodal joint generative model of brain data. In: International conference on information processing in medical imaging, pp 17–29
Ledig C, Heckemann RA, Hammers A, Lopez JC, Newcombe VF, Makropoulos A, Lötjönen J, Menon DK, Rueckert D (2015) Robust whole-brain segmentation: application to traumatic brain injury. Med Image Anal 21(1):40–58
Rekik I, Allassonniére S, Carpenter TK, Wardlaw JM (2012) Medical image analysis methods in mr/ct-imaged acute-subacute ischemic stroke lesion: segmentation, prediction and insights into dynamic evolution simulation models. A critical appraisal. Neuroimage Clin 1(1):164–178
Hevia-Montiel N, Jiménez-Alaniz JR, Medina-Bańuelos V, Yáñez-Suárez O, Rosso C, Samson Y, Baillet S (2007) Robust nonparametric segmentation of infarct lesion from diffusion-weighted mr images. In: International conference of the IEEE engineering in medicine & biology society, pp 2102–2105
Liu J, Li M, Wang J, Wu F, Liu T, Pan Y (2014) A survey of mri-based brain tumor segmentation methods. Tsinghua Sci Technol 19(6):578–595
Rajini NH, Bhavani R (2013) Computer aided detection of ischemic stroke using segmentation and texture features. Measurement 46(6):1865–1874
Griffis JC, Allendorfer JB, Szaflarski JP (2016) Voxel-based Gaussian naïve Bayes classification of ischemic stroke lesions in individual t1-weighted MRI scans. J Neurosci Methods 257:97–108
Chen L, Bentley P, Rueckert D (2017) Fully automatic acute ischemic lesion segmentation in dwi using convolutional neural networks. Neuroimage Clin 15:633–643
Zhang R, Zhao L, Lou W, Abrigo JM, Mok VC, Chu WC, Wang D, Shi L (2018) Automatic segmentation of acute ischemic stroke from dwi using 3d fully convolutional densenets. IEEE Trans Med Imaging 37(9):2149–2160
Liu Z, Cao C, Ding S, Han T, Wu H, Liu S (2018) Towards clinical diagnosis: automated stroke lesion segmentation on multimodal mr image using convolutional neural network, arXiv preprint arXiv:1803.05848, pp 1–20
Havaei M, Dutil F, Pal C, Larochelle H, Jodoin PM (2015) A convolutional neural network approach to brain tumor segmentation. In: International Workshop on Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries, pp 195–208
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2016) Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med Image Anal 36:61–78
Forbes F, Doyle S, Garcia-Lorenzo D, Barillot C, Dojat M (2010) Adaptive weighted fusion of multiple mr sequences for brain lesion segmentation. In: IEEE international conference on biomedical imaging, pp 69–72
Seghier ML, Ramlackhansingh A, Crinion J, Leff AP, Price CJ (2008) Lesion identification using unified segmentation-normalisation models and fuzzy clustering. Neuroimage 41(4):1253–1266
James JR, Yoder KK, Osuntokun O, Kalnin A, Bruno A, Morris ED (2006) A supervised method for calculating perfusion/diffusion mismatch volume in acute ischemic stroke. Comput Biol Med 36(11):1268–1287
Maier O, Wilms M, von der Gablentz J, Krämer UM, Münte TF, Handels H (2015) Extra tree forests for sub-acute ischemic stroke lesion segmentation in mr sequences. J Neurosci Methods 240:89–100
Maier O, Menze BH, von der Gablentz J, Häni L, Heinrich MP, Liebrand M, Winzeck S, Basit A, Bentley P, Chen L (2017) Isles 2015-a public evaluation benchmark for ischemic stroke lesion segmentation from multispectral mri. Med Image Anal 35:250–269
Bauer S, Nolte LP, Reyes M (2012) Skull-stripping for tumor-bearing brain images, arXiv preprint arXiv:1204.0357, 2012
Beumer D, Rozeman AD, Nijeholt GJL, Brouwer PA, Jenniskens SFM, Algra A, Boiten J, Schonewille W, Oostenbrugge RJV, Dippel DWJ (2016) The effect of age on outcome after intra-arterial treatment in acute ischemic stroke: a mr clean pretrial study. BMC Neurol 16(1):1–7
González RG, Schaefer PW, Buonanno FS, Schwamm LH, Budzik RF, Rordorf G, Wang B, Sorensen AG, Koroshetz WJ (1999) Diffusion-weighted mr imaging: diagnostic accuracy in patients imaged within 6 hours of stroke symptom onset. Radiology 210(1):155–162
Moseley M, Kucharczyk J, Mintorovitch J, Cohen Y, Kurhanewicz J, Derugin N, Asgari H, Norman D (1990) Diffusion-weighted mr imaging of acute stroke: correlation with t2-weighted and magnetic susceptibility-enhanced mr imaging in cats. Am J Neuroradiol 11(3):423–429
Moseley M, Cohen Y, Mintorovitch J, Chileuitt L, Shimizu H, Kucharczyk J, Wendland M, Weinstein P (1990) Early detection of regional cerebral ischemia in cats: comparison of diffusion-and t2-weighted mri and spectroscopy. Magn Reson Med 14(2):330–346
Sorensen AG, Copen WA, Ostergaard L, Buonanno FS, Gonzalez RG, Rordorf G, Rosen BR, Schwamm LH, Weisskoff RM, Koroshetz WJ (1999) Hyperacute stroke: simultaneous measurement of relative cerebral blood volume, relative cerebral blood flow, and mean tissue transit time. Radiology 210(2):519–527
Collins DL, Neelin P, Peters TM, Evans AC (1994) Automatic 3d intersubject registration of mr volumetric data in standardized talairach space. J Comput Assist Tomogr 18(2):192–205
Jenkinson M, Pechaud M, Smith S (2005) Bet2: Mr-based estimation of brain, skull and scalp surfaces. In: Eleventh annual meeting of the organization for human brain mapping, vol 17, p 167
Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Išgum I (2016) Automatic segmentation of mr brain images with a convolutional neural network. IEEE Trans Med Imaging 35(5):1252–1261
Bauer S, Wagner M, Seiler A, Hattingen E, Deichmann R, Nóth U, Singer OC (2014) Quantitative t2’-mapping in acute ischemic stroke. Stroke 45(11):3280–3286
Mohammadi-Nejad AR, Hossein-Zadeh GA, Soltanian-Zadeh H (2017) Structured and sparse canonical correlation analysis as a brain-wide multi-modal data fusion approach. IEEE Trans Med Imaging 36(7):1438–1448
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) Imagenet: a large-scale hierarchical image database. In: IEEE conference on computer vision and pattern recognition, pp 248–255
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118
Chatfield K, Simonyan K, Vedaldi A, Zisserman A (2014) Return of the devil in the details: delving deep into convolutional nets. arXiv:1405.3531
Dosovitskiy A, Fischer P, Springenberg J, Riedmiller M, Brox T (2016) Discriminative unsupervised feature learning with exemplar convolutional neural networks. IEEE Trans Pattern Anal Mach Intell 38(9):1734–1747
Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, Hulsbergen-Van De Kaa C, Bult P, Van Ginneken B, Van Der Laak J (2016) Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6:26286
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in mri images. IEEE Trans Med Imaging 35(5):1240–1251
He K, Sun J (2015) Convolutional neural networks at constrained time cost. In: IEEE conference on computer vision and pattern recognition, pp 5353–5360
Srivastava RK, Greff K, Schmidhuber J (2015) Highway networks, arXiv preprint arXiv:1505.00387
Nair V, Hinton GE (2010) Rectified linear units improve restricted boltzmann machines. In: International conference on international conference on machine learning, pp 807–814
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241
He K, Zhang X, Ren S, Sun J (2016) Identity mappings in deep residual networks. In: European conference on computer vision, pp 630–645
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation applied to handwritten zip code recognition. Neural Comput 1(4):541–551
Milletari F, Navab N, Ahmadi S (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: Fourth international conference on 3D Vision, pp 565–571
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: IEEE conference on computer vision and pattern recognition, pp 3431–3440
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. In: Deep learning and data labeling for medical applications, pp 179–187
Drozdzal M, Chartrand G, Vorontsov E, Shakeri M, Di LJ, Tang A, Romero A, Bengio Y, Pal C, Kadoury S (2017) Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal 44:1–13
Dice LR (1945) Measures of the amount of ecologic association between species. J Ecol 26(3):297–302
Huttenlocher DP, Klanderman GA, Rucklidge WA (1993) Comparing images using the Hausdorff distance. IEEE Trans Pattern Anal Mach Intell 15(9):850–863
Saad NM, Abu-Bakar S, Muda S, Mokji M, Salahuddin L (2011) Brain lesion segmentation of diffusion-weighted mri using gray level co-occurrence matrix. In: IEEE international conference on imaging systems and techniques, pp 284–289
Benjamini Y (1988) Opening the box of a boxplot. Am Stat 42(4):257–262
He K, Zhang X, Ren S, Sun J (2015) Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: IEEE international conference on computer vision, pp 1026–1034
Maas AL, Hannun AY, Ng AY (2013) Rectifier nonlinearities improve neural network acoustic models. Int Conf Mach Learn 30:3
Zhu X, Zhang S, Hu R, Zhu Y, Song J (2017) Local and global structure preservation for robust unsupervised spectral feature selection. IEEE Trans Knowl Data Eng 30(3):517–529
Liu J, Li M, Lan W, Wu F-X, Pan Y, Wang J (2018) Classification of alzheimer’s disease using whole brain hierarchical network. IEEE/ACM Trans Comput Biol Bioinf 15(2):624–632
Zhu Y, Zhu X, Kim M, Yan J, Kaufer D, Wu G (2018) Dynamic hyper-graph inference framework for computer assisted diagnosis of neurodegenerative diseases. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2018.2868086
Kong Y, Gao J, Xu Y, Pan Y, Wang J, Liu J (2019) Classification of autism spectrum disorder by combining brain connectivity and deep neural network classifier. Neurocomputing 324(9):63–68
Liu J, Wang X, Zhang X, Pan Y, Wang X, Wang J (2018) Mmm: classification of schizophrenia using multi-modality multi-atlas feature representation and multi-kernel learning. Multimed Tools Appl 77(22):29651–29667
Acknowledgements
The authors would like to thank Chunlei Liu, an Associate Professor in the Department of Electrical Engineering and Computer Sciences, UC Berkeley, for his valuable comments on this study. The work described in this paper was supported by the National Natural Science Foundation of China under Grant Nos. 61772557, 61772552, 61622213, 61728211; the 111 Project (No.B18059); the Hunan Provincial Science and Technology Program (2018WK4001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, L., Chen, S., Zhang, F. et al. Deep convolutional neural network for automatically segmenting acute ischemic stroke lesion in multi-modality MRI. Neural Comput & Applic 32, 6545–6558 (2020). https://doi.org/10.1007/s00521-019-04096-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-019-04096-x