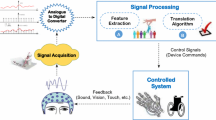
Abstract
This paper studies the state-of-the-art classification techniques for electroencephalogram (EEG) signals. Fuzzy Functions Support Vector Classifier, Improved Fuzzy Functions Support Vector Classifier and a novel technique that has been designed by utilizing Particle Swarm Optimization and Radial Basis Function Networks (PSO-RBFN) have been studied. The classification performances of the techniques are compared on standard EEG datasets that are publicly available and used by brain–computer interface (BCI) researchers. In addition to the standard EEG datasets, the proposed classifier is also tested on non-EEG datasets for thorough comparison. Within the scope of this study, several data clustering algorithms such as Fuzzy C-means, K-means and PSO clustering algorithms are studied and their clustering performances on the same datasets are compared. The results show that PSO-RBFN might reach the classification performance of state-of-the art classifiers and might be a better alternative technique in the classification of EEG signals for real-time application. This has been demonstrated by implementing the proposed classifier in a real-time BCI application for a mobile robot control.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
With the improvement in biomedical technology and machine learning, the analysis of human bio-potential signals has become an important research field. Researchers have been studying to understand and classify human bio-signals in order to better help people with various facilities as in developing assistive technologies for the disabled or obtaining more accurate diagnosis of diseases [1]. Among the human bio-potentials, electroencephalogram (EEG) brain signals have been widely studied because of their clinical importance for non-invasive approaches of detecting neurological abnormalities and a broad range of applications for brain–computer interface technologies [2, 3].
EEG signals are electrical action potential signals recorded on the scalp, generated by the firing of many neurons in the brain. It has been known that specific tasks in the human body are controlled by particular parts of the brain [4]. The firing of the neurons results in generating intensive neurological electrical activity enough to be measured by the non-invasive electrodes placed on specific parts of the brain. The recorded EEG signals are within 1–100 μV amplitude range and may contain useful information to decipher human thoughts or intents. This information can be converted into control inputs for various real-time systems such as BCI-based assistive devices or detection systems for brain-related abnormal activities [5].
Difficult characteristics of EEG signals, such as poor signal-to-noise ratio compared to other human bio-potentials, require employment of robust classification algorithms in order to achieve reasonable classification performance. Therefore, designing efficient classification algorithms has been an important goal and highly attractive area for the research community [2, 3].
Classification of EEG signals mainly consists of three components. The first component is preprocessing the EEG signals. Preprocessing is the phase of preparing the data in order to remove undesired components that might constitute outliers in the signal such as eye blink artifacts for EEG signal. The second component is feature extraction where various pattern recognition and signal processing techniques (e.g., PCA, FFT or wavelet transforms) might be employed in order to extract meaningful features that might be enough for classification [6, 7]. The last component is employing a machine learning technique in order to classify the EEG signal utilizing the data generated in the feature extraction step.
Our study focuses on an application of state-of-the-art classifiers on EEG datasets while designing an additional efficient classification technique. This proposed technique utilizes Radial Basis Function networks (RBFN) classifier, which is a simple one-layer feed-forward type of Neural Networks. As it will be introduced later in more detail, clustering algorithms might be employed in order to find the parameters of RBFN classifier and they might highly affect the performance of the classifier [8–12]. Thus, in order to discover the effects of better clustering on RBFN classifier, the performances of several clustering algorithms such as K-means, Fuzzy C-means and Particle Swarm Optimization (PSO) clustering are studied with comparison. Finally, the classification performance of the RBFN classifier together with the efficient clustering algorithm is compared with recent classifiers such as FFSVC and IFFSVC.
The following section introduces EEG classification and the offline datasets used throughout this study as well as the employed feature extraction technique for the EEG signals. The third section gives brief information about the studied classification techniques. Section 4 introduces the clustering techniques and their performance comparisons. The experimental results related to the classifiers are discussed in Sect. 5. Finally, a real-time BCI implementation of the proposed classification algorithm is presented in Sect. 6 followed by the conclusions.
2 EEG collection, datasets and feature extraction
In this section, the collection methods of EEG signals such as the electrode placement system and the electrodes used to collect the signals are introduced first. The details of the offline datasets used in this study are explained. Then, the feature extraction method employed to process the EEG signals is presented.
2.1 Collection of EEG signals and the EEG datasets
In order to collect EEG signals, the electrodes are placed according to a standard system called International 10–20 system as shown in Fig. 1. In this system, each electrode is named with a letter to identify the lobe and a number to identify the hemispheric location. Odd numbers refer to the left hemisphere, and even numbers refer to the right hemisphere of the scalp. On the basis of the specific mental tasks, the signal coming from several individual electrodes might be of an interest.
For example, Sensorimotor Cortex part of the brain where the human motor actions are controlled in general corresponds to the locations on the central lobe of the brain [13].
The offline datasets used in this study are two-class datasets obtained from BCI Competition IIIb and IV2b [14, 15] named as ×11, O3VR, S4b (Competition IIIb) and B0101T, B0102T (Competition IV2b). For ×11, O3VR and S4b, the data are collected from different subjects at multiple sessions including several trials each. On the other hand, B0101T and B0102T include the data that belong to the same subject but collected through multiple training sessions on two different days within 2 weeks. The electrode positions placed on the scalp of subjects for each dataset are C3, C4 and Cz as shown in Fig. 1. Note that these electrode positions are within the Sensorimotor Cortex Area of the brain. The EEG signal in dataset IIIb is sampled with 125 Hz and preprocessed by filtering between 0.5 and 30 Hz [14]. On the other hand, the EEG signal in datasets of IV2b has a sampling frequency of 250 Hz and filtered between 0.5 and 100 Hz [15].
The data are collected according to the cue-based screening paradigm where subjects are shown motor imaginary pictures as cues and data are recorded accordingly. Timing intervals for collection of datasets from Competition IIIb are shown in Fig. 2. One trial contains 8 s of recording. Shortly after a fixation cross is displayed on the screen, a short cue beep is generated as a warning to the subject, indicating that one of the two visual cue images will be displayed. The cue images displayed to the subjects on the screen are either left arrow or right arrow indicating left thinking or right thinking. After the visual cue is displayed, through a virtual reality experiment, a feedback is given to the subject such as moving a ball to the left or right.
Unlike the IIIb datasets, B0101T and B0102T datasets are collected without any feedback given to the subject but with the similar cue-based scheme. The detailed timing schematic for the collection of IV2b datasets is given in Fig. 3.
2.2 The feature extraction method for EEG datasets
An important phenomenon with the EEG signals is known to be Mu rhythm or Event-Related Desynchronization (ERD) [14]. This event creates a characteristic attenuation in the power of EEG signal in certain frequency ranges due to motor action preparation by the Sensorimotor Cortex Area of the brain. Although this rhythm is observed in the planning stage of physical movements, it has been discovered by Pfurtscheller [14] that it might also appear when the human is shown a visual stimulus as a mental preparation of physical actions. During this mental preparation, an instant power decrease in the EEG signal occurs and the power of the signal increases again. Therefore, the re-synchronization of the EEG signal is called Event-Related Synchronization (ERS).
Considering the nature of ERD and ERS phenomena, in order to extract features from the EEG signal, band power (BP) values of the signal are extracted within the subject’s most reactive frequency ranges, alpha [8–11] Hz and beta [11–29] Hz, as suggested in [14] and [16]. Band power values are obtained by band-pass filtering the signal within specific frequency ranges, squaring it and taking the average. After generating BP values, mean band power value of the signal is taken within the time interval starting from the visual cue demonstration and until the end of visual feedback or imaginary period. As a result of feature extraction, a four-dimensional feature vector is obtained for each trial such as [C3α, C3β, C4α and C4β] where C3α represents the band power feature, extracted from C3 within the alpha band frequency range.
3 The classification techniques
In this study, three state-of-the-art classifiers are implemented and tested on the same standard datasets and compared with each other. Among these, Fuzzy Support Vector Classifier (FFSVC) and Improved Support Vector Classifier (IFFSVC) are implemented because of their excellent performances on the regular datasets that exceed the well-known Support Vector Classifier [17–19]. In addition, it has been reported by the studies in [2] and [16] that Fuzzy Classifiers have been rarely used in EEG classification research, which suggests a great research potential. In addition to these two classifiers, an RBFN- and PSO-based classification algorithm is proposed. Note that this study is also important from the stand point that the FFSVC and IFFSVC techniques have been specifically implemented and tested for the first time in classifying EEG signals.
3.1 Fuzzy functions support vector classifier (FFSVC)
The Fuzzy Functions Support Vector Classifier is a new classifier design proposed by Celikyilmaz et al. [17]. It combines the Fuzzy C-means clustering (FCM) algorithm with any classification methods (Support Vector Classifier in our case) to design a new classifier. This classifier approach captures the hidden partitions in the dataset using FCM clustering and applies one classifier for each partition found by the clustering method. The membership values found by FCM clustering are augmented with the original training feature set. This helps classification by increasing the dimensionality of the input space so that the data might more likely become linearly separable. Since each classifier helps estimating a part of the decision boundary, they might be considered as a fuzzy model that represents conventional linguistic fuzzy IF–THEN rules in Fuzzy Inference Systems [17].
The general structure describing FFSVC classification technique is shown in Fig. 4.
Fuzzy functions support vector classifier schematic [13]
Let each data vector in the dataset includes nv number of features and k = 1, …, nv. The data vector with nv features can be represented as x = [x 1, …, x nv ]. For each fuzzy classifier, corresponding membership value, μ i , which is obtained by Fuzzy C-means clustering algorithm, is included as additional features to the data vector for each cluster i and i = 1, …, c. Therefore, for the special case of single data vector, the augmented vector can be represented as ϕ i = [x 1, …, x nv , …, μ i ]. These augmented vectors are created for each data vector in the dataset according to the results of clustering algorithm. The fuzzy classifier function layer includes a classification method selected as Support Vector Classifiers (SVC). The output of each fuzzy classifier is the probabilistic outputs of SVC, generated by Platt’s probability method [20]. In the output layer, probability outputs are weighted with the membership values and summed up in order to find a crisp probability output in the sense of defuzzification in Fuzzy Inference Systems (FIS) [17, 18]. The final result is thresholded in order to generate true class labels.
In employing this classifier, there are three parameters that need to be adjusted m, c and Creg. The variables m and c come from the standard FCM clustering algorithm where m is the fuzzification constant that determines the degree of overlapping in clusters and the variable c is the number of clusters that the dataset will be partitioned. The constant Creg is the regularization constant for SVC that is used to adjust the position of the separation plane. In employing SVC for this paper, for all datasets, the radius of the Gaussian kernel in SVC is taken as 1/nv where nv is the number of features in an input vector and the other three parameters are found by grid search as suggested in [17].
Improved Fuzzy Functions Support Vector Classification (IFFSVC) is a newer technique that optimizes the membership values obtained with standard FCM clustering in order to be a better predictor for the data. This has been done by adding additional error term to the objective function of standard FCM algorithm and modifying the membership update formula accordingly [19].
3.2 Particle swarm optimization-based radial basis function networks classifier (PSO-RBFN)
Radial Basis Function Networks (RBFN) is a type of feed-forward Neural Network, which consists of three layers: input layer, hidden layer and output layer as shown schematically in Fig. 5. The input layer contains n-dimensional feature vectors entering the network. The hidden layer is composed of radially symmetric Gaussian kernel functions as shown in (1)
where i = 1, 2, 3, …, m and m being the number of kernels, x i represents the ith kernel center in the hidden layer and x k represents the kth feature vector in the dataset. Values of ϕ i are calculated for each data vector together with kernel centers, which might be determined by any clustering technique [21]. Note that, the closer x k is to x i , the higher the influence it will have in the hidden layer outputs.
By the help of the hidden layer, feature vectors, which are in R n, are mapped into a higher dimension, R m, so that the data can more likely become linearly separable according to the Cover’s theorem [21].
The outputs of hidden layer units are connected to the output layer by weighted links. The output node of RBFN is a linear summation as described in (2)
where W j represents the weights of links between hidden and output layers and y i represents the output value generated by the network for the ith data vector. Training of the network includes finding the appropriate weights between the hidden layer and the output layer that will provide appropriate mapping for the data vector from input space to the output space.
Let ϕ ij represent the hidden layer value for the jth kernel and the ith data vector in the dataset, and the output layer value can be written in the matrix form as follows in (3)
where N represents the number of data vectors in the dataset. The weights can be calculated using the pseudo-inverse between output layer matrix, Y, and hidden layer matrix, ϕ, as stated in (4).
Using the pseudo-inverse of matrix ϕ provides minimization of the mean square error between the actual value of the output layer and the estimated value generated by the RBF network [21]. During the testing phase of the data with RBFN, y i outputs are found using the weights obtained in the training. These outputs are thresholded at the end in order to generate binary class label outputs.
In employing RBFN for a classification problem, finding the appropriate centers for kernel functions has critical importance on the generalization capability of the classifier. Therefore, several clustering algorithms are widely used in the literature to supervise the cluster centers [8–12]. In this study, PSO clustering algorithm is used to determine the cluster centers for the hidden layer. The inspiration for employing PSO clustering as a clustering technique is the promising result of our earlier work [22]. In [22], and the superior properties of PSO are examined over well-known clustering algorithms such as K-means [23] and Fuzzy C-means [24] using six different datasets. The following section briefly summarizes our experimental results obtained in [22] for the clustering algorithms.
4 Comparisons of clustering techniques
This section presents the three clustering techniques: Fuzzy C-means [24], K-means [23] and Particle Swarm Optimization (PSO) clustering [25]. Their clustering performances are compared using the standard datasets. In the literature, the clustering performance of PSO has been shown to be superior compared to other clustering techniques [22, 25].
Particle Swarm Optimization clustering is based on standard PSO technique where each particle in the swarm is a potential solution to the optimization problem (possible cluster centers for clustering), which is determined by a fitness function to be minimized or maximized. As the algorithm iterates, each particle is allowed to update its position in the search space evaluating its own fitness and the fitness of the neighboring particles. The algorithm is terminated when the specified maximum number of iterations is reached or there is no improvement in the global best solution of the swarm. The particle that has the best fitness at the end of the last iteration is selected as the solution. For each particle, x k ∈ R nv, the velocity and position of particles are updated based on (5) and (6), respectively.
where v k (n) = current velocity of particle k, x k (n) = current position of particle k, p k = best position of particle k, g(n) = current global best position of the swarm, β c = cognitive weight, β s = social weight and r = random number (0,1).
In recent years, there have been many improvements and modifications to the original PSO since it has been proposed by [26]. One of these modifications is called the Constriction PSO [27], which is also utilized in this study and proven to give better results compared to other types of modifications such as Inertia PSO [28].
The Constriction PSO is a way to guarantee the convergence in the system through the assignment of eigenvalues with a constriction coefficient determined by cognitive and social acceleration coefficients. The velocity and position updates for this type of PSO algorithm are shown in (7), (8).
In (7) and (8), the χ coefficient is called the constriction constant that is used to guarantee the population stability and is given by (9) below
In this equation, ϕ = β c + β s and is calculated to be ϕ > 4 in order to guarantee the convergence [27]. The constant K allows the user to tweak the degree of convergence. Setting K = 1 provides a slower convergence but a more explorative searching as shown by [27]. Therefore, K = 1 is also used for our implementation.
On the basis of the experimental results presented by both [27] and [28] and to guarantee the stability, the above parameters are selected as: χ = 0.729 and β c = U(1, 4) (uniformly distributed random number between 1 and 4) and β s = 4.1 − β c . Note that, the greater the constriction factor than 0.729, the faster the algorithm converges, however, the probability of convergence also decreases.
The three clustering algorithms are compared in their abilities to minimize the quantization error, which is a measure for clustering performance [25, 29] defined by the formula in (10).
In (10), |C j | represents the number of data vectors belonging to the cluster j that is the frequency of that cluster, m j represents the center of cluster j, z p is the pth data vector in the dataset and N c is the number of clusters that the data will be partitioned. The quantization error in (10) is a measure of how close the center locations are to the data members in each cluster. Minimizing the quantization error makes the intra-cluster distances decrease and inter-cluster distances increase, therefore results in more compact clustering [25].
Table 1 compares the clustering algorithms and their performances by presenting the mean final quantization errors and standard deviations after 10 runs. During these runs, all of the algorithms are started from the same random initial cluster centers. In PSO clustering, all of the particles are initialized to the same center locations with other clustering algorithms. The PSO clustering is run with 20 particles and 50 maximum iterations, while the K-means and FCM are run with 1,000 maximum iterations, in order to have the same number of fitness evaluations, and are completed in each method for the fair comparison.
The datasets used in this table are standard datasets used in the literature [30]. As it can be seen from the results in Table 1, PSO clustering is better than other methods in terms of obtaining smaller quantization error and getting closer to the global solution. In order to better demonstrate the results of Table 1, quantization error versus number of fitness evaluations plot for E. coli dataset is plotted in Fig. 6 for a single run.
The plot shows that although Fuzzy C-means and K-means clustering algorithms converge a lot faster than PSO clustering, these two algorithms can easily get stuck in local minima and do not show any progress no matter how long the algorithms are run. However, since the PSO algorithm works in a more explorative way, as the algorithm is kept running, it continues and finds better minima than the other two algorithms. Note that, since the PSO clustering is run with 20 particles and 50 maximum iterations, the number of fitness evaluations that have been performed in each iteration is 20. Therefore, the number 1,000 in Fig. 6 corresponds to the result of 50 iterations of the PSO clustering algorithm. On the other hand, since each iteration of FCM and K-means corresponds to one objective function evaluation, 1,000 fitness evaluations indicate that both algorithms have run 1,000 steps.
It should be noted that although the PSO algorithm has superior capabilities in doing better clustering, it comes with a trade-off. PSO clustering converges slower than K-means and FCM based on the number of fitness evaluations. This is because PSO has explorative abilities, whereas K-means and FCM have only exploitative abilities. Thus, they have faster convergence but most likely end up in a local optimum.
The same convergence plots are generated for standard EEG datasets obtained from BCI Competition IIIb and IV2b [14, 15] where the details of data collection and feature extraction methods are introduced in Sect. 2. Analyzing the results in the plots, the PSO clustering resulted in smaller quantization error for all datasets. Thus, it may improve the performance of classifier, which employs clustering (Fig. 7).
5 Experimental results
In this section, the classification results of three classifiers are presented on the two types of datasets namely EEG and non-EEG datasets as introduced earlier and used in the clustering section.
The FFSVC and IFFSVC classifiers are implemented utilizing LIBSVM libraries in Matlab [31]. The parameter optimizations of these classifiers are performed as suggested in [17, 19]. A grid search is performed in order to obtain the combination of parameters that gives the best cross-validation performance within the following ranges for Creg = [27, 26, …, 2−4], m = [1.3, 1.4, …, 2.4] and c = [2, 3, …, 8].
As introduced in Sect. 3, the parameter optimization of RBFN includes finding hidden layer kernel centers and the optimum number of hidden layer units that might generalize the data well enough. During the parameter optimization of the RBFN, the number of hidden layer units is searched exhaustively by starting from the same number of feature dimension of the dataset and increasing until the two times of dimension. The cross-validation is done according to the average result of both training and validation performances. This is done to avoid overtraining the classifier. While employing PSO-RBFN, the PSO algorithm is run 100 iterations with the number of particles adjusted according to the formula in (11) as suggested by [25]
where D = Number of features*Number of clusters.
The initial cluster centers are selected randomly and different for each particle in the swarm. On the other hand, the initial cluster centers of FCM clustering are taken the same as one of the particles in the swarm so that it is guaranteed FCM clustering starts at the same location with one of the particles in the swarm. After the hidden layer centers are found by each clustering algorithm, the standard deviation values for the Gaussian kernels are searched within the range of 0.1, 0.2, 0.3, …, 10, 20, 30, 40, 50 by comparing the cross-validation performances. The number of hidden layer units and the standard deviation that give the highest average training and validation performance is chosen for testing of the classifier on the new data. The fitness function of PSO is selected as quantization error, and fitness values of the particles are evaluated according to this error during PSO runs. Table 2 includes the dataset names and the ratios that the datasets are partitioned as training, validation and test data.
The three classifiers are run 10 times, and the classification performances are recorded. Table 3 includes the average classification, standard deviation and the maximum classification performance reached during algorithm runs for the PSO-RBFN classifier.
Similarly, Table 4 includes the same type of results for RBFN classifier but in this case FCM algorithm is used to find the centers of the hidden layer units for the classifier. This is done in order to compare the effect of clustering algorithms on the classification performance of RBFN classifier.
When the results in both tables are analyzed, it can be seen that the PSO-RBFN algorithm does significantly better than FCM-RBFN classifier for some of the datasets. For instance, for B0102T dataset, each PSO-RBFN run has exceeded the FCM-RBFN performance. Another promising result of PSO clustering is that it helps RBFN to reach the maximum classification performance during data runs. For Ionosphere dataset, it is observed that the performance of PSO-RBFN has reached to 100% where it was significantly better than any other FCM-RBFN runs. This is because of the more explorative searching abilities of PSO algorithm as compared to FCM, bringing more variation to the clustering that can reach the maximum performance over all.
Finally, the results obtained from FFSVC and IFFSVC classifiers are presented in Tables 5 and 6.
Analyzing the overall results, it can be concluded that PSO-RBFN classifier may compete with the state-of-the-art FFSVC and IFFSVC classifiers in the standard datasets. When the average maximum results are compared, PSO-RBFN performance exceeds FFSVC peformance. Although the average performance values of FFSVC and IFFSVC exceed the PSO-RBFN results, using the PSO-RBFN classifier for EEG classification applications might be still advantageous because of their low computational complexity and their extensibility. Both of the FFSVC and IFFSVC classifiers are designed to be two-class classifiers. However, with a slight modification, such as increasing the number of nodes in the output layer of RBFN, the PSO-RBFN classifier can be easily extended for a multi-class classification problem.
In addition, the simplicity that comes with implementing and training the RBFN classifier and the less computational complexity might still make the PSO-RBFN more preferable to the FFSVC and IFFSVC classifiers.
6 A real-time BCI application
The commercial Emotiv Epoc ® EEG data acquisition headsets available in our research laboratory are used for a real-time application of PSO-RBFN classifier in a BCI system. The classified outputs are used for control inputs of an external device such as a mobile robot called Hexapod as shown in Fig. 8a.
The Emotiv Epoc headsets are originally created in order to provide an innovative way of game control using brain–computer interface (BCI) technology [32]. With the research edition of the headsets, the raw EEG data can be accessed and processed utilizing the Software Development Kit libraries, which are written in C++. In order to utilize easy and efficient signal processing built-in functions of Matlab software and provide an easy development environment for other BCI researchers, a software interface has been written in Matlab integrating C++ dynamic linking libraries (dll) of the headsets so that they can be programmed by only using Matlab.
A schematic of the electrode positions of the headsets is given in Fig. 8b.
In this study, the electrodes FC5, F3, F4, FC6, O1 and O2 are used as they relate to the Sensorimotor Cortex (FC5, F3, F4 and FC6) and Visual Cortex areas (O1 and O2) of the brain. These areas are known to manage planning, control and execution of motor actions of the body [3, 14].
In order to provide a visual stimulus to the subject for the left or right thinking similar to the standard EEG datasets used in this paper, the EmoCube ® stand-alone application is used. The EmoCube works as stand-alone executable server software written by Emotiv that accepts UDP packets in order to move a 3-D cube to the several locations in the screen within the main frame. The neutral position of the EmoCube is shown in Fig. 9.
In order to perform the training and testing for the real-time system, a graphical user interface has been created utilizing the Matlab Graphical User Interface (GUI) design environment. The design of the system GUI is shown in Fig. 10.
In Fig. 10, left and right training buttons are used to supervise the subject by providing visual stimuli via UDP packets sent to the EmoCube server. When the left or right button is pressed, the EmoCube moves to the left or right from the center neutral position and returns back to its neutral position again. The process that the cube moves to the leftmost or rightmost positions takes 8 s. With the start of cube movement, data acquisition is started, and as the cube is moving, the raw EEG data collected from the headset electrodes are stored into a buffer after filtering the data between the frequency ranges of 0.5–30 Hz. The mean band power features within alpha and beta bands are extracted from the single trial by ignoring the initial 2 s. Finally, these features are saved into a global feature buffer.
When the Train PSO-RBF button is pressed, the PSO-RBFN classifier is trained with the training samples collected during the first session. This has been determined according to the cross-validation results of the classifier. The weight matrix and the hidden layer parameters of the RBFN classifier are also found in this step. Finally, by pressing the Real-Time Test button, the raw EEG data are continuously buffered with the intervals of 6 s and decisions are made by feeding the extracted band power features to the classifier as test vectors.
The decisions generated by the PSO-RBFN classifier in real time are classified either left or right thinking and sent to the mobile robot wirelessly to move the robot to the right or left. It should be noted that the wrong classified directions are not used to re-train the system. The system is initially trained with 450 samples that are collected on different multiple training sessions from the same subject. The training samples are selected randomly from the buffer by separating the 50% of the samples as training and the remaining as for the cross-validation reasons. It has been observed that the cross-validation performance of the system has reached up to 65% during the training, and the subject was able to move the robot in 7 correct directions out of 10 pre-determined directions, suggesting that a feasible real-time control of a robot with EEG signals is possible. Although the number of experiments is considered to be good enough to see the general performance of the algorithm, the number of runs can be increased in order to get a more accurate statistical measurement.
7 Conclusion
This paper provides a study on the applications of various classification algorithms including the proposed PSO-RBFN classifier. In order to observe the effect of clustering algorithms on the performance of RBFN classifier, three well-known clustering algorithms are compared with each other by their abilities to minimize the quantization error. It can be concluded that better clustering obtained with PSO algorithm helps RBFN to improve its classification performance.
When the PSO-RBFN classifier is compared with the other two classifiers, the PSO-RBFN might reach or exceed the performance level of FFSVC and IFFSVC for most of the datasets. The extensibility of PSO-RBFN classifier for a multi-class problem and its simple implementation and training for the classification tasks make the PSO-RBFN classifier preferable for a real-time EEG classification application. To support this conclusion, the proposed classifier is successfully implemented for a real-time brain–computer interface application in order to control a mobile robot with commercial EEG headsets.
References
Dickhaus H, Heinrich H (1996) Classifiying biosignals with wavelet networks, a method for noninvasive diagnosis. IEEE Eng Med Biol Mag 15:103–111
Lotte F, Congedo M, Lecuyer A, Lamarche F, Arnaldi B (2007) A review of classification algorithms for eeg-based brain-computer interfaces. J Neural Eng 4:R1–R13
Sanei S, Chambers J (2007) EEG signal processing. Wiley-Interscience, NJ
Okamoto M, Dan H, Sakamoto K, Takeo K (2004) Three-dimensional probabilistic anatomical cranio-cerebral correlation via the international 10–20 system oriented for transcranial functional brain mapping. Neuroimage 21:99–111
Takahashi K, Nakauke T, Hashimoto M (2005) Remarks on hands-free manipulation using bio-potential signals. In: Proceedings of IEEE international conference on systems man and cybernetics, pp 2965–2970
Subasi S (2007) EEG signal classification using wavelet feature extraction and a mixture of expert model. Expert Syst Appl 32(4):1084–1093
Polat K, Gunes S (2008) Artificial immune recognition system with fuzzy resource allocation mechanism classifier, principal component analysis and FFT method based new hybrid automated identification system for classification of EEG signals. Expert Syst Appl 34(3):2039–2048
Uykan Z, Guzelis C, Celebi ME, Koivo HN (2000) Analysis of input-output clustering for determining centers of RBFN. IEEE Trans Neural Netw 11(4):851–858
Pedrycz W (1998) Conditional fuzzy clustering in the design of radial basis function networks. IEEE Trans Neural Netw 9(4):601–612
Staiano A, Tagliaferri R, Pedrycz W (2006) Improving RBF networks performance in regression tasks by means of a supervized fuzzy clustering. Neurocomputing 69(15):1570–1581
Hongyang L, He J (2009) The application of dynamic K-means clustering algorithm in the center selection of RBF neural networks. In: 3rd international conference on genetic and evolutionary computing vol 177, no. 23, pp 488–491
Scholkopf B, Sung KK, Burges CJC, Girosi F, Niyogi P, Poggio T, Vapnik V (1997) Comparing support vector machines with Gaussian kernels to radial basis function classifiers. IEEE Trans Signal Process 45(11):2758–2765
Drongelen WV (2007) Signal processing for neuroscietistis. Elselvier, Burlington
Pfurtscheller G, Neuper C (2001) Motor imagery and direct brain-computer communication. Proc IEEE 89:1123–1134
Blankertz B, Dornhege G, Krauledat M, Müller KR, Curio G (2007) The non-invasive Berlin brain-computer interface: fast acquisition of effective performance in untrained subjects. Neuroimage 37(2):539–550
Zhong M, Lotte F, Girolami M, Lecuyer A (2008) Classifying EEG for brain computer interfaces using gaussian processes. Pattern Recogn Lett 29(3):354–359
Celikyilmaz A, Turksen IB (2009) A new classifier design with fuzzy functions. In: Proceedings of the 11th international conference on rough sets, fuzzy sets, data mining and granular computing, pp 1123–1134
Celikyilmaz A, Turksen IB (2007) Fuzzy functions with support vector machines. Inf Sci 177(23):5163–5177
Celikyilmaz A, Turksen IB, Aktas R, Doganay MM (2009) Increasing accuracy of two-class pattern recognition with enhanced fuzzy functions. Expert Syst Appl 36(2P1):1337–1354
Platt J (2000) Probabilistic outputs for support vector machines and comparison to regularized likelihood methods. MIT Press, Cambridge
Haykin S (ed) (2008) Neural networks: a comprehensive foundation, 3rd edn. Prentice Hall, Upper Saddle River
Johnson R, Sahin F (2009) Particle swarm optimization methods for data clustering. In: Proceedings of the soft computing, computing with words and perceptions in system analysis, decision and control, 2009. ICSCCW 2009. Fifth International Conference, pp 1–6
Alpaydin E (2004) Introduction to machine learning. The MIT Press, Cambridge
Pal NR, Pal K, Bezdek JC (1997) A mixed C-means clustering model. In: Proceedings of the sixth IEEE international conference on fuzzy systems
Van der Merwe DW, Engelbrecht AP (2003) Data clustering using particle swarm optimization. In: Proceedings of IEEE congress on evolutionary computation, pp 215–220
Kennedy J, Eberhart RC (1995) Particle swarm optimization. In: Proceedings of the IEEE international conference on neural networks, vol IV, pp 1942–1948
Clerc M, Kennedy J (2002) The particle swarm—explosion, stability, and convergence in a multidimensional complex space. IEEE Trans Evol Comput 6(1):58–73
Eberhart RC, Shi Y (2000) Comparing inertia weights and constriction factors in particle swarm. In: Proceedings of the 2000 congress on evolutionary computation, pp 84–88
Cox E (2005) Fuzzy modelling and genetic algorithms for data mining and exploration. Elseveir, San Francisco
Asuncion A, Newman DJ (2007) UCI machine learning repository. University of California, Irvine, school of information and computer sciences
LIBSVM: a library for support vector machines [online]. Avaiable: http://www.csie.ntu.edu.tw/~cjlin/libsvm. Accessed June 2010
Emotiv: brain computer interface technology [online]. Avaiable: http://www.emotiv.com. Accessed June 2010
Acknowledgments
The authors would like to thank the reviewers for their valuable comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cinar, E., Sahin, F. New classification techniques for electroencephalogram (EEG) signals and a real-time EEG control of a robot. Neural Comput & Applic 22, 29–39 (2013). https://doi.org/10.1007/s00521-011-0744-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-011-0744-x