Abstract
An Electrocardiogram or ECG is an electrical recording of the heart and is used in the investigation of heart disease. This ECG can be classified as normal and abnormal signals. The classification of the ECG signals is presently performed with the support vector machine. The generalization performance of the SVM classifier is not sufficient for the correct classification of ECG signals. To overcome this problem, the ELM classifier is used which works by searching for the best value of the parameters that tune its discriminant function and upstream by looking for the best subset of features that feed the classifier. The experiments were conducted on the ECG data from the Physionet arrhythmia database to classify five kinds of abnormal waveforms and normal beats. In this paper, a thorough experimental study was done to show the superiority of the generalization capability of the Extreme Learning Machine (ELM) that is presented and compared with support vector machine (SVM) approach in the automatic classification of ECG beats. In particular, the sensitivity of the ELM classifier is tested and that is compared with SVM combined with two classifiers, and they are the k-nearest Neighbor Classifier and the radial basis function neural network classifier, with respect to the curse of dimensionality and the number of available training beats. The obtained results clearly confirm the superiority of the ELM approach as compared with traditional classifiers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
ECG is a technique which captures transthoracic interpretation of the electrical activity of the heart overtime and externally recorded by skin electrodes. The electrical potential generated by electrical activity in cardiac tissue is measured on the surface of the human body. Current flow, in the form of ions, signals contraction of cardiac muscle fibers leading to the heart’s pumping action. It is a non-persistent recording produced by an electrocardiographic device. The recognition and classification of the ECG beats is a very important task in the coronary intensive unit, where the classification of the ECG beats is essential tool for the diagnosis. ECG offers cardiologists with useful information about the rhythm and functioning of the heart. Therefore, its analysis represents an efficient way to detect and treat different kinds of cardiac diseases Up to now, many algorithms have been developed for the recognition and classification of ECG signal. Some of them use time and some use frequency domain for depiction. Based on that many specific attributes are defined, allowing the recognition between the beats belonging to different pathological classes. The ECG waveforms may be different for the same patient to such extent that they are unlike each other and at the same time alike for different types of beats [1]. Artificial Neural Network (ANN) and fuzzy-based techniques were also employed to exploit their natural ability in pattern recognition task for successful classification of ECG beats [2].
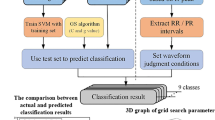
In this paper, the approach to ECG beat classification presented thorough experimental exploration of the ELM capabilities for ECG classification. Further, the performances of the ELM approach in terms of classification accuracy are evaluated: (1) by automatically detecting the best discriminating features from the whole considered feature space and (2) by solving the model selection issue. Unlike traditional feature selection methods, where the user has to specify the number of desired features, the proposed system gives a method for extraction of features called as “feature detection”. Feature selection and feature detection have the common characteristic of searching for the best discriminative features. The latter, however, has the advantage of determining their number automatically. In other words, feature detection does not require the desired number of most discriminative features from the user apriori. The detection process is implemented through AR Modeling framework that exploits a criterion intrinsically related to ELM classifier properties. This framework is formulated in such a way that it also solves the model selection issue, i.e., to estimate the best values of the ELM classifier parameters, which are the regularization and kernel parameters.
The rest of the paper is organized as follows. The AR method for ECG feature extraction, the basic mathematical formulation of SVMs for solving binary and multiclass classification problems, and the working methodology of ELM are given in Sect. 3. The experimental results obtained on ECG data from the Massachusetts Institute of Technology–Beth Israel Hospital (MIT–BIH) arrhythmia database [9] are reported in Sect. 4. Finally, conclusions are drawn in Sect. 5.
2 Literature survey
In the literature survey, several methods have been proposed for the automatic classification of ECG signals. Among the most recently published works are those presented as follows
Khadra et al. [3] proposed a high-order spectral analysis technique for quantitative analysis and classification of cardiac arrhythmias. The algorithm is based upon bispectral analysis techniques. Autoregressive model is used to estimate the bispectrum, and the frequency support of the bispectrum is extracted as a quantitative measure to classify a trial and ventricular tachyarrhythmias. A significant difference in the parameter values for different arrhythmias is observed in the result. Furthermore, the bicoherency spectrum shows different bicoherency values for normal and tachycardia patients. The bicoherency indicates in particular that phase coupling decreases as arrhythmia kicks in. The simplicity of the classification parameter and the obtained sensitivity and specificity of the classification scheme reveal the importance of higher-order spectral analysis in the classification of life-threatening arrhythmias.
de Chazal et al. [4] investigate the design of an efficient system for recognition of the premature ventricular contraction from the normal beats and other heart diseases. This system comprises three main modules: denoising module, feature extraction module, and classifier module. In the denoising module, it has proposed the stationary wavelet transform for noise reduction of the electrocardiogram signals. In the feature extraction of the ECG module, a proper combination of the morphological-based features and timing interval-based features is proposed. As the classifier, a number of supervised classifiers are investigated; they are as follows: a number of multilayer perceptron neural networks with different number of layers and training algorithms, support vector machines with different kernel types, radial basis function, and probabilistic neural networks. Also, for comparison the proposed features, the author has considered the wavelet-based features. It has done comprehensive simulations to achieve a high-efficient system for ECG beat classification from 12 files obtained from the MIT–BIH arrhythmia database. Simulation results show that best results are achieved about 97.14% for classification of ECG beats.
Andreao et al. [5] proposed a novel-embedded mobile ECG reasoning system that integrates ECG signal reasoning and RF identification together to monitor an elderly patient. As a result, this proposed method has a good accuracy in heart beat recognition and enables continuous monitoring and identification of the elderly patient when alone. Moreover, in order to examine and validate this proposed system, the author proposes a managerial research model to test whether it can be implemented in a medical organization. The results prove that the mobility, usability, and performance of author’s proposed system have impacts on the user’s attitude, and there is a significant positive relation between the user’s attitude and the intent to use the proposed system.
Mitra et al. [6] put forth a three stage technique for detection of premature ventricular contraction (PVC) from normal beats and other heart diseases. This method includes a feature extraction module, a denoising module, and a classification module. In the first module, the author investigates the application of stationary wavelet transform (SWT) for noise reduction of the electrocardiogram (ECG) signals. The feature extraction module finds out 10 ECG morphological features and one timing interval feature. Then, a number of MLP (multilayer perceptron) neural networks with different number of layers and nine training algorithms are designed. The network’s performance for speed of convergence and accuracy classifications are evaluated for seven files from the MIT–BIH arrhythmia database. Among the various training algorithms, the resilient back-propagation (RP) algorithm illustrated the best convergence rate and the Levenberg–Marquardt (LM) algorithm achieved the best overall detection accuracy.
Sheng-Wu Xiong et al. [8] proposed in their paper that fuzzy support vector machines based on fuzzy c-means clustering. They apply the fuzzy c-means clustering method to each class of the training set. At the time of the clustering with a suitable fuzziness parameter q, the much important samples, such as support vectors, become the cluster centers, respectively.
Siew et al. [7] give an idea on ELM. In this paper, they presented Extreme Learning Machine (ELM) for Single-hidden Layer Feed-forward Neural-networks (SLFNs), which randomly chooses hidden nodes and analytically determines the output weights of SLFNs. The ELM avoids problems like improper learning rate, local minima, and overfitting commonly faced by iterative learning methods and completes the training very fast. The author has evaluated the multicategory classification performance of ELM on five different datasets related to bioinformatics namely, the Breast Cancer Wisconsin dataset, the Pima Diabetes dataset, the Heart-Statlog dataset, the Hepatitis dataset, and the Hypothyroid dataset. A detailed analysis of different activation functions with unreliable number of neurons is also carried out, which concludes that Algebraic Sigmoid function outperforms all other activation functions on these datasets. The evaluation results indicate that ELM provides better classification accuracy with reduced training time and implementation complexity compared with earlier implemented models. Emanet [23] presented an ECG beat classification by using discrete wavelet transform and Random Forest algorithm. Wen et al. [24] use GreyART network for ECG beat classification.
Nazmy et al. [21] present a novel ECG classification approach. This is an intelligent diagnosis system using hybrid approach of Adaptive Neuro-Fuzzy Inference System (ANFIS) model for classification of Electrocardiogram (ECG) signals. Feature extraction using Independent Component Analysis (ICA) and Power spectrum, together with the RR interval then serve as input feature vector, this feature was used as input of ANFIS classifiers. Six types of ECG signals are normal sinus rhythm (NSR), premature ventricular contraction (PVC), atrial premature contraction (APC), Ventricular Tachycardia (VT), Ventricular Fibrillation (VF), and Supraventricular Tachycardia (SVT). The proposed ANFIS model combined the neural network adaptive capabilities and the fuzzy inference system. The results indicate a high level of efficient of tools used with an accuracy level of more than 97%. This section presented the literature survey on the previous ECG classification techniques. Sabry et al. [22] proposed a third-order cumulant signature matching technique for non-invasive fetal heart beat identification.
3 Methodology
3.1 Feature extraction
Automatic ECG beat recognition and classification [20] are performed in the part either by the neural network or by the other recognition systems relying in various features, time domain representation, extracted from the ECG beat [2], or the measure of energy in a band of frequencies in the spectrum (frequency domain representation) [10]. Since these features are very at risk to variations of ECG morphology and the temporal characteristics of ECG, it is difficult to distinguish one from the other on the basis of the time waveform or frequency representation. In this paper, three different classes of feature set are used belonging to the isolated ECG beats including third-order cumulant, autoregressive model parameters, and the variance of discrete wavelet transform detail coefficients for the different scales (1–6 scales).
3.1.1 Wavelet transformation
Physiologies used for diagnosis are frequently characterized by a non-stationary time behavior. For such patterns, time and frequency representations are desirable. The frequency characteristics in addition to the temporal behavior can be described with respect to uncertainty principle. The wavelet transform can represent signals in different resolutions by dilating and compressing its basis functions. While the dilated functions adapt to slow wave activity, the compressed functions capture fast activity and sharp spikes. The most favorable choice of types of wavelet functions for preprocessing is problem dependent. In this paper, Daubechies wavelet function (db5) which is called compactly supported orthonormal wavelets [11]. By making discretization the scaling factor and position factor, the DWT is obtained. For orthonormal wavelet transform, x(n), the discrete signal can be expanded into the scaling function at j level as follows:
where D j,k represents the detailed signal at j level. Note that j controls the dilation or contraction of the scale function Φ(t), k denotes the position of the wavelet function Ψ(t), and n represents the sample number of the x(n). Here, n∈Z represents the set of integers. The frequency spectrum of the signal is classified into high frequency and low frequency for wavelet decomposition as the band increases (j = 1, …, 6). Wavelet transform is a two-dimensional timescale processing method for non-stationary signals with adequate scale values and shifting in time [12].
Multiresolution decomposition can efficiently provide simultaneous characteristics, in term of the representation of the signal at multiple resolutions corresponding to different timescales. Feature vectors are constructed by the normalized variances of detail coefficients of the DWT which belongs to the related scales.
3.1.2 Higher-order statistics and AR modeling
The main problem in automatic ECG beat recognition and classification is that related features are very susceptible to variations of ECG morphology and temporal characteristics of ECG. In the study [1] the set of original QRS complexes typical for six types of arrhytmia taken from the MIT/BIH arrhytmia database, there is a great variations of signal among the same type of beats belonging to the same type of arrhytmia. Therefore, in order to solve such problem, the author will rely on the statistical features of the ECG beats. In this paper for this aim, third-order cumulant has been taken into account, which can be determined (for zero mean signals) as follows
where E represents the expectation operator, and k, l, and m are the time lags. In this paper, third-order cumulant of selected ECG beats is used. Normalized ten points represent the cumulant evenly distributed with in the range of 25 lags. Each succeeding samples of a signal as a linear combination of previous samples, that is, as the output of an all-pole IIR filter is modeled by linear prediction. This process locates the coefficients of an nth order autoregressive linear process that models the time series x as
where x represents the real input time series (a vector), and n is the order of the denominator polynomial a(z). In the block processing, autocorrelation method is one of the modeling methods of all-pole modeling to find the linear prediction coefficients. This method is as well called as the maximum entropy method (MEM) of spectral analysis.
3.2 Support vector machines
SVM is usually used for classification tasks introduced by Vapnik [13]. For binary classification, SVM is used to find an optimal separating hyperplane (OSH), which generates a maximum margin between two categories of data. To construct an OSH, SVM maps data into a higher-dimensional feature space. SVM performs this non-linear mapping by using a kernel function. Then, SVM constructs a linear OSH between two categories of data in the higher feature space. Data vectors which are nearest to the OSH in the higher feature space are called support vectors (SVs) and contain all information required for classification. In brief, the theory of SVM is as follows [13].
Consider training set \( D = \left\{ {(x_{j} ,y_{i} )} \right\}_{i = 1}^{L} \)with each input n i x ∈ R n and an associated output y i ∈{−1, +1}. Each input x is firstly mapped into a higher dimension feature space F, by z = φ(x) via a non-linear mapping φ: R n → F. When data are linearly non-separable in F, there exists a vector w ∈ F and a scalar b which define the separating hyperplane as follows:
Here, ξ(≥0) are called slack variable. The hyperplane that optimally separates the data in F is one that
where C is called regularization parameter that determines the trade-off between maximum margin and minimum classification error. By constructing a Lagrangian, the optimal hyperplane according to (7) may be shown as the solution of
where α1,…..,α L is the non-negative Lagrangian multipliers. The data points i x that correspond to αi > 0 are SVs. The weight vector w is then given by
For any test vector x ∈ Rn, the classification output is then given by
To build an SVM classifier, a kernel function and its parameters need to be chosen. So far, no analytical or empirical studies have established the superiority of one kernel over another conclusively. The kernel K(·,·) must satisfy the condition stated in Mercer’s theorem so as to correspond to some type of inner product in the transformed (higher) dimensional feature space Φ(X) [14]. A typical example kernels used is represented by the following Gaussian function:
where γ is a parameter which is inversely proportional to the width of the Gaussian kernel.
As described before, SVMs are intrinsically binary classifiers. But, the classification of ECG signals often involves the simultaneous discrimination of numerous information classes. In order to face this issue, a number of multiclass classification strategies can be adopted [15, 16]. The most popular ones are the one-against-all (OAA) and the one-against-one (OAO) strategies. The former involves a reduced number of binary decompositions (and thus, of SVMs), which are, however, more complex. The latter requires a shorter training time, but may incur conflicts between classes due to the nature of the score function used for decision. Both strategies generally lead to similar results in terms of classification accuracy. In this paper, the OAA strategy is considered. Briefly, this strategy is based on the following procedure. Let Ω = {ω 1 , ω 2 ,…,ω T } be the set of T possible labels (information classes) associated with the ECG beats that desired to classify. First, an ensemble of T (parallel) SVM classifiers is trained. Each classifier aimed at solving a binary classification problem defined by the discrimination between one information class ωi(i = 1, 2,…,T) against all others (i.e., Ω − {ωi}). Then, in the classification phase, the new rule is used to decide which label to assign to each beat which is “winner-takes-all” rule. This represents that the winning class is the one that corresponds to the SVM classifier of the ensemble that shows the highest output (discriminant function value).
3.3 Extreme learning machine
A new learning algorithm called the ELM for SLFNs supervised batch learning. The output of an SLFN with ~N hidden nodes (additive or RBF nodes) can be represented by
where a i and b i are the learning parameters of hidden nodes, and βi is the weight connecting the ith hidden node to the output node. G(a i , b i , X) is the output of the ith hidden node with respect to the input x. For the additive hidden node with the activation function g(x): R → R (e.g., sigmoid or threshold), G(a i , b i , X) is given by
where a i represents the weight vector connecting the input layer to the ith hidden node, and bi is the bias of the ith hidden node. a i ·x denotes the inner product of vectors ai and x in Rn. For an RBF hidden node with an activation function g(x): R → R (e.g., Gaussian), G(a i , b i , X) is given by
where a i and b i are the ith RBF node’s center and impact factor. R + indicates the set of all positive real values. The RBF network is a special case of the SLFN with RBF nodes in its hidden layer. Each RBF node has its own centroid and impact factor and output of it is given by a radially symmetric function of the distance between the input and the center.
In the learning algorithms, it uses a finite number of input–output samples for training. Here, N arbitrary distinct samples are considered (x i , t i ) ∈ R n x R m, where x i is an n × 1 input vector and t i is an m × 1 target vector. If an SLFN with \( \tilde{N} \) hidden nodes can approximate N samples with zero error, it then implies that there exist β i , a i , and b i such that
Equation () can be written compactly as
where
H is called the hidden layer output matrix of the network [15]; the ith column of H is the ith hidden node’s output vector with respect to inputs x 1, x 2,…, x N and the jth row of H is the output vector of the hidden layer with respect to input x j .
In real applications, the number of hidden nodes, \( \tilde{N}, \) will always be less than the number of training samples, N, and, hence, the training error cannot be made exactly zero but can approach a non-zero training error. The hidden node parameters ai and bi (input weights and biases or centers and impact factors) of SLFNs need not be tuned during training and may simply be assigned with random values according to any continuous sampling distribution. Equation 18 then becomes a linear system, and the output weights are estimated as
where \( H\dag \) the Moore–Penrose is generalized inverse [15] of the hidden layer output matrix H. The ELM algorithm, which consists of only three steps, can then be summarized as
ELM Algorithm: Given a training set \( \aleph = \{ (X_{i} ,t_{i} )|X_{i} \in R^{n} ,t_{i} \in R^{m} ,\quad i = 1, \ldots ,N\} \) activation function g(x), and hidden node number \( \tilde{N}, \)
-
1.
Assign random hidden nodes by randomly generating parameters (a i , b i ) according to any continuous sampling distribution, i = 1,…., \( \tilde{N} \)
-
2.
Calculate the hidden layer output matrix H.
-
3.
Calculate the output weight β: \( \tilde{\beta } = H\dag T \)
The universal approximation capability of ELM has been analyzed by Huang et al. [16] using an incremental method and it shows that single SLFNs with randomly generated additive or RBF nodes with a wide range of activation functions can universally approximate any continuous target functions in any compact subset of the Euclidean space Rn. \( g\left( x \right) = {\frac{1}{{1 + e^{ - \lambda x} }}} \) is the sigmoidal function used as activation function in ELM.
4 Experimental results
4.1 Dataset description
The experiment conducted on the basis of ECG data from the Physionet database [9]. In particular, the considered beats refer to the following classes: normal sinus rhythm (N), atrial premature beat (A), ventricular premature beat (V), right bundle branch block (RB), left bundle branch block (LB), and paced beat (/). The beats were selected from the recordings of 20 patients, which correspond to the following files: 100, 102, 104, 105, 106, 107, 118, 119, 200, 201, 202, 203, 205, 208, 209, 212, 213, 214, 215, and 217. In order to feed the classification process, in this paper, the two following kinds of features are adopted: (1) ECG morphology features and (2) three ECG temporal features, i.e., the QRS complex duration, the RR interval (the time span between two consecutive R points representing the distance between the QRS peaks of the present and previous beats), and the RR interval averaged over the ten last beats [4]. In order to extract these features, first the QRS detection is performed and ECG wave boundary recognition tasks by means of the well-known ecgpuwave software available on [17]. Then, after extracting the three temporal features of interest, normalized to the same periodic length, the duration of the segmented ECG cycles according to the procedure is reported in [18]. To this purpose, the mean beat period was chosen as the normalized periodic length, which was represented by 300 uniformly distributed samples. Consequently, the total number of morphology and temporal features equals 303 for each beat.
In order to obtain reliable assessments of the classification accuracy of the investigated classifiers, in all the following experiments, three different trials are performed, each with a new set of randomly selected training beats, while the test set was kept unchanged. The results of these three trials obtained on the test set were thus averaged. The detailed numbers of training and test beats are reported for each class in Table 1. Classification performance was evaluated in terms of four measures, which are: (1) the overall accuracy (OA), which is the percentage of correctly classified beats among all the beats considered (independently of the classes they belong to); (2) the accuracy of each class that is the percentage of correctly classified beats among the beats of the considered class; (3) the average accuracy (AA), which is the average over the classification accuracies obtained for the different classes; (4) the McNemar’s test that gives the statistical significance of differences between the accuracies achieved by the different classification approaches. This test is based on the standardized normal test statistic [19]
where Z ij measures the pairwise statistical significance of the difference between the accuracies of the ith and jth classifiers. f ij stands for the number of beats classified correctly and wrongly by the ith and jth classifiers, respectively. Accordingly, f ij and f ji are the counts of classified beats on which the considered ith and jth classifiers disagree. At the commonly used 5% level of significance, the difference of accuracies between the ith and jth classifiers is said statistically significant if |Zij | > 1.96.
4.2 Experimental scheme
The proposed experimental framework was performed around the following five main experiments. The first experiment aimed at assessing the effectiveness of the SVM approach in classifying ECG signals directly in the whole original hyperdimensional feature space (i.e., by means of all the 303 available features). The total number of training beats was fixed to 500, as reported in Table 1. For comparison purpose, two other reference non-parametric classification approaches are implemented, namely, the kNN and the RBF neural network classifiers. In the second experiment, it was desired to explore the behavior of the SVM classifier (compared to the two reference classifiers) when integrated within a standard classification scheme based on an AR feature reduction. In particular, the number of features was varied from 10 to 50 with a step of 10 so as to test this classifier in small as well as high-dimensional feature subspaces. The third experimental part had for objective to assess the capability of the proposed ELM classification system to boost further the accuracy of the SVM classifier.
The fourth experiment was devoted to analyze the generalization capability of the SVM, the kNN, and the RBF classifiers with and without feature reduction, and of the ELM classification system by decreasing/increasing the number of available training beats. This analysis was done through two experimental scenarios, which consisted in passing from 500 to 250 and 750 training beats, respectively. Finally, in the fifth experiment, the sensitivity of the ELM classification system is analyzed.
4.3 Experimental settings
In the experiments, the non-linear SVM is considered based on the popular Gaussian kernel (referred to as SVM-RBF or simply SVM). The related parameters γ and C for this kernel were varied in the arbitrarily fixed ranges [10−3, 200] and [10−3, 2] so as to cover high and small regularization of the classification model, and fat as well as thin kernels, respectively. In addition, for comparison purpose, in the first experiment, the SVM classifier with two other kernels is implemented, which are the linear and the polynomial kernels, leading thus to two other SVM classifiers termed as SVM-linear and SVM-poly, respectively.
The polynomial kernel’s degree d was varied in the range [2, 5] in order to span polynomials with low and high flexibility. The K value and the number of hidden nodes (h) of the kNN and the RBF classifiers were tuned in the arbitrarily fixed intervals [1, 15] and [10, 60], respectively. The other RBF parameters, which include the center and the width of each RBF (kernel), were computed by applying the K-means clustering algorithm separately to each class.
In this experiment, the SVM classifier is trained based on the Gaussian kernel, which proved in the previous experiments to be the most appropriate kernel for ECG signal classification, in feature subspaces of various dimensionalities. The desired number of features varied from 10 to 50 with a step of 10, namely, from small- to high-dimensional feature subspaces. Feature reduction was achieved by the traditional AR modeling, commonly used in ECG signal classification. In particular, it can be seen that for all feature subspace dimensionalities except the lowest (i.e., 10 features), the ELM classifier maintains a clear superiority over the other two. Its best accuracy was found using a feature subspace made up of the first 30 components. The corresponding OA and AA accuracies were 89.74 and 89.78%, respectively. Comparing these results with those achieved with the SVM classifier based on the Gaussian kernel in the original feature space (i.e., without feature reduction), a slight increase of 1.98% in terms of OA and 2.30% in terms of AA was obtained, which is represented in Table 2. From this experiment, three observations can be made: (1) the SVM classifier shows a relatively low sensitivity to the curse of dimensionality as compared with the kNN and the RBF classifiers (2) the SVM classifier still preserve its superiority when integrated in a feature reduction-based classification scheme; and (3) though the SVM performs well in the whole original feature space, its accuracy can still be improved provided that a subspace of higher generalization capability can be found.
The Fig. 1 gives the comparison of the accuracy of classifying the ECG signals by using SVM-rbf and ELM. This shows that ELM gives much better accuracy for all datasets given as input in which RB dataset achieves the maximum accuracy of 97.69%. The Fig. 2 gives the comparison of the accuracy of classifying the ECG signals by using SVM-kNN and ELM. This shows that ELM gives much better accuracy for all datasets given as input. The Fig. 3 gives the comparison of the accuracy of classifying the ECG signals by using SVM-poly and ELM. This shows that ELM gives much better accuracy for all datasets given as input.
As described before, the proposed ELM classification system aimed at enhancing the SVM classification process from two different viewpoints: (1) by automatically detecting a feature subspace of higher generalization capability in order to deal in a more effective way with the curse of dimensionality, instead of reducing the dimension of the original feature space basing on reduction algorithm and (2) by passing from an empirical tuning of the value of the two SVM parameters to their automatic optimization. This experiment is aimed at assessing the effectiveness of this methodological enhancement. To this purpose, the ELM classifier is applied to the available training beats.
At convergence of the optimization process, the ELM classifier’s accuracy on the test samples assessed. The achieved overall and average accuracies were 89.74 and 89.78% corresponding to substantial accuracy gains are higher as compared with SVM combined with various kernel functions. Its worst class accuracy was obtained for normal beat (N) (89.69%), while that of the SVM and the ELM classifiers was for ventricular premature beats (V) as they were (81.48%) and (85.18%), respectively. This shows the capability of the ELM classifier to reduce the gap between the worst and the best class accuracies while keeping OA at a high level.
Table 3 shows the number of features detected automatically to discriminate each class from the others. The average number of features required by the ELM classifier is 47, while the minimum and maximum numbers of features were obtained for the ventricular premature (V) and normal (N) classes with 32 and 68 features, respectively.
5 Conclusion
In this paper, a novel ECG beat classification system using ELM is proposed and applied to MIT/BIH data base. The wavelet transforms variance and AR model parameters have been used for the features selection. From the obtained experimental results, it can be strongly recommended the use of the ELM approach for classifying ECG signals on account of their superior generalization capability as compared with traditional classification techniques. This capability generally provides them with higher classification accuracies and a lower sensitivity to the curse of dimensionality. The results confirm that the ELM classification system substantially boosts the generalization capability achievable with the SVM classifier and its robustness against the problem of limited training beat availability, which may characterize pathologies of rare occurrence. Another advantage of the ELM approach can be found in its high sparseness, which is explained by the fact that the adopted optimization criterion is based on minimizing the number of SVs. It can also be seen that ELM accomplishes better and more balanced classification for individual categories as well in very less training time comparative to SVM. In future, some advanced neural network techniques can be used to train the ELM classifier, and it may enhance the classification accuracy of the ECG and reduce the training time.
References
Osowski S, Linh TH (2001) ECG beat recognition using fuzzy hybrid neural network. IEEE Trans Biomed Eng 48(11):1265–1271
Hu YH, Palreddy S, Tompkins W (1997) A patient adaptable ECG beat classifier using a mixture of experts approach. IEEE Trans Biomed Eng 44(9):891–900
Khadra L, Al-Fahoum AS, Binajjaj S (2005) A quantitative analysis approach for cardiac arrhythmia classification using higher order spectral techniques. IEEE Trans Biomed Eng 52(11):1840–1845
de Chazal F, Reilly RB (2006) A patient adapting heart beat classifier using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 53(12):2535–2543
Andreao RV, Dorizzi B, Boudy J (2006) ECG signal analysis through hidden Markov models. IEEE Trans Biomed Eng 53(8):1541–1549
Mitra S, Mitra M, Chaudhuri BB (2006) A rough set-based inference engine for ECG classification. IEEE Trans Instrum Meas 55(6):2198–2206
Siew C-K, Huang G-B (2005) Extreme learning machine with randomly assigned RBF Kernels. Int’l J Inf Technol 11(1):16–24
Xiong S-W, Liu H-B, Niu X-X (2005) Fuzzy support vector machines based on FCM clustering. In: Proceedings of 2005 international conference on machine learning and cybernetics, August 2005, vol. 5, pp 2608–2613
Mark R, Moody G (1997) MIT-BIH Arrhythmia Database 1997 [Online]. Available http://ecg.mit.edu/dbinfo.html
Minami K, Nakajima H, Toyoshima T (1999) Real-time discrimination of ventricular tachyarrhytmia with Fourier transform neural network. IEEE Trans Biomed Eng 46:179–185
Daubechies I (1998) Orthonormal bases of compactly wavelets. Commun Pure Appl Math (41):909–996
Thakor NV (1993) Multiresolution wavelet analysis of evoked potentials. IEEE Trans Biomed Eng 40(11):1085–1093
Vapnik VN (1995) The nature of statistical learning theory. Springer, New York, 187 pp
Vapnik VN (1998) Statistical learning theory. Wiley, New York
Melgani F, Bruzzone L (2004) Classification of hyperspectral remote sensing images with support vector machine. IEEE Trans Geosci Remote Sens 42(8):1778–1790
Hsu C-W, Lin C-J (2002) A comparison of methods formulticlass support vector machines. IEEE Trans Neural Netw 13(2):415–425
Wei JJ, Chang CJ, Shou NK, Jan GJ (2001) ECG data compression using truncated singular value decomposition. IEEE Trans Biomed Eng 5(4):290–299
Agresti A (2002) Categorical data analysis, 2nd edn. Wiley, New York
Park H-S, Woo S-M, Kim Y-S, Kang B-J, Ban S-W (2009) ECG pattern classification based on generic feature extraction. In: Proceedings of the 3rd WSEAS international conference on circuits, systems, signal and telecommunications (CISST’2009), pp 21–25
Nazmy TM, El-Messiry H, Al-bokhity B (2009) Adaptive neuro-fuzzy inference system for classification of ECG signals. J Theor Appl Inf Technol
Sabry-Rizk M, Zgallai W, Hardiman P, O’Riordan J (1997) Third-order cumulant signature matching technique for non-invasive fetal heart beat identification. In: IEEE international conference on acoustics, speech, and signal processing, vol. 5, pp 3781–3784
Emanet N (2009) ECG beat classification by using discrete wavelet transform and Random Forest algorithm. In: Fifth international conference on soft computing, computing with words and perceptions in system analysis, decision and control, pp 1–4
Wen C, Yeh MF, Chang KC (2007) ECG beat classification using GreyART network. Signal Process IET 1(1):19–28
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Karpagachelvi, S., Arthanari, M. & Sivakumar, M. Classification of electrocardiogram signals with support vector machines and extreme learning machine. Neural Comput & Applic 21, 1331–1339 (2012). https://doi.org/10.1007/s00521-011-0572-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-011-0572-z