Abstract
Purpose
We recently reported frequent detection of antibiotic-resistant bacteria on the oral mucosa during the period of hematopoietic cell transplantation (HCT) and suggested an association between oral mucositis and antibiotic-resistant bacterial infection. Methicillin-resistant Staphylococcus spp. were frequently detected, and the oral cavity may be a reservoir of the gene mediating methicillin resistance, mecA. Here, we examined the frequency of mecA carriers in patients undergoing HCT.
Methods
Fifty-nine patients (male (M) = 37, female (F) = 22, 47.3 ± 11.0 years) receiving HCT were enrolled in this study. Buccal swab samples were obtained four times from day −7 to day +20 (once/week), and mecA was detected by PCR. Fifty-two subjects without systemic disease, who completed dental treatment, especially periodontal treatment (M = 21, F = 31, 55.4 ± 14.2 years), were also enrolled as controls and checked for mecA on the oral mucosa.
Results
Seventy-six percent (45/59) of the HCT patients carried mecA at least once in the study period (days −7 to +20), while no control subjects had mecA. The frequency of mecA carriers was 19.2 % from days −7 to −1, while it was significantly increased on days +7 to +13 and +14 to +20, with frequencies of 60.9 and 63.2 %, respectively (P < 0.01, ANOVA).
Conclusions
mecA was detected in oral mucosa of patients undergoing HCT. The high detection frequency of staphylococci resistant to penicillin and beta-lactams in our recent report was supported.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral mucositis is one of the most common symptomatic complications associated with chemotherapy, especially hematopoietic stem cell transplantation (HCT) [12, 13]. Severe mucositis is associated with not only intolerable pain but also the possible risk of systemic bacteremia. Oral mucositis is a significant cause of suffering and morbidity in patients receiving myeloablative chemotherapy [1].
Severe mucositis is associated with a risk of systemic infection related to bacteremia. Our recent studies showed that not only normal oral flora but also opportunistic bacteria appear on the oral mucosa. Bacterial substitution of mainly coagulase-negative staphylococci (CoNS) for streptococci occurred frequently on the oral buccal mucosa after HCT, and other bacterial species not usually found in the normal flora were also identified [9]. We reported that multidrug-resistant opportunistic bacteria appearing in the gingiva may be involved in fatal sepsis [11]. Furthermore, many antibiotic-resistant bacteria were detected in the oral cavity after HCT, especially during the period in which the severity of oral mucositis reaches its peak [10]. CoNS and Staphylococcus aureus with penicillin and beta-lactam resistance were detected [10].
Penicillin and beta-lactam resistance among staphylococci is mediated by point mutations in penicillin-binding proteins (PBPs) [2]. Methicillin resistance in staphylococci is mediated by the mecA gene complex, which is located on a unique molecular vector called the staphylococcal chromosome cassette (SCCmec) [3]. SCCmecs carry mobility genes and integrate in a site-specific manner into a highly conserved locus in the Staphylococcus chromosome.
The present study was performed to determine the distribution of oral mucosal bacteria with mecA in patients undergoing hematopoietic cell transplantation.
Materials and methods
Subjects
Fifty-nine patients (male (M) = 37, female (F) = 22, 47.3 ± 11.0 years) receiving HCT at Okayama University Hospital from 2011 to 2012 were enrolled in this study. The diseases of these patients are shown in Table 1. Autologous HCT, conventional allogeneic HCT, and reduced-intensity stem cell transplantation (RIST) were administered to 12 (M = 8, F = 4, 56.8 ± 9.5 years), 13 (M = 9, F = 4, 42.4 ± 10.6 years), and 34 (M = 20, F = 14, 54.6 ± 11.4 years) patients, respectively. Fifty-two patients without systemic diseases, who visited the Department of Periodontics, Okayama University Hospital (M = 21, F = 31, 55.4 ± 14.2 years), were also enrolled as controls. A total of 111 subjects were enrolled in the study. Informed consent for examination of oral bacteria was obtained from each subject, and the Ethics Committee of Okayama University Graduate School of Medicine, Dentistry, and Pharmaceutical Sciences approved this study (no. 457).
Oral management
Intensive oral care was performed for all HCT subjects as in our previous report [10]. All HCT subjects received were referred to dentists, and necessary dental treatment was completed before HCT. All subjects received instruction regarding self-management of oral hygiene, tooth brushing after every meal and before going to bed, and oral rinsing with normal saline solution every 3 h during the day was also indicated. Nurses, dental hygienists, and dentists performed these oral managements in cases in which the patient’s condition was poor. No antibiotic rinses were used.
All control subjects were in maintenance phase after completion of dental treatment, especially for periodontal treatment. Therefore, their oral hygiene was well maintained. Subjects who had systemic diseases were excluded. All subjects confirmed that they had not received antibiotic treatment for at least 3 months prior to enrolling in the study.
General infection control for HCT subjects
All HCT patients were isolated in a room equipped with a laminar airflow system and received trimethoprim-sulfamethoxazole as prophylaxis against Pneumocystis carinii. Fluoroquinolone for prophylaxis against bacterial infection and fluconazole for prophylaxis against fungal infection were administered orally. Prophylaxis against herpes virus infection with acyclovir was also given. Neutropenic fever was managed according to the guidelines of Hughes et al. [5]. Briefly, empirical antibiotic therapy was administered promptly in all neutropenic patients at the onset of fever and in afebrile patients who were neutropenic but who had signs or symptoms compatible with infection. A fourth-generation cephalosporin (e.g., cefepime) or carbapenem (e.g., meropenem) was administered intravenously as empirical antibiotic therapy. G-CSF (lenograstim 5 μg/kg/day or filgrastim 300 μg/m2) was given intravenously for 60 min starting on day 1 or day 5 and was continued until the absolute neutrophil count exceeded 500/μL.
Collection of bacterial samples
Microbial samples were obtained from HCT patients about 2 h after breakfast by swabbing from the whole surface of the buccal mucosa regardless of whether mucositis was observed. Collection of bacterial samples was performed four times (days −7 to −1, days 0 to +6, days +7 to +13, and days +14 to +20) for each patient (a total of 236 times in 59 patients). However, samples could not be collected 27 times because of the patents’ conditions. A total of 209 samples were subjected to mecA detection procedures.
Microbial samples were also obtained from control subjects once just after a checkup and before any dental intervention at our hospital. Thus, the dental treatment on the checkup day could not affect the results of this study. A total of 52 samples from 52 control subjects were subjected to mecA detection procedures.
Detection of mecA
-
1.
Bacterial DNA extraction
Cotton swab samples were suspended in 1 mL of PBS(−) (Gibco BRL, Grand Island, NY). Aliquots of 500 μL from each suspension were transferred into new tubes and pelleted. Pelleted samples were resuspended in 200 μL of InstaGene matrix (Bio-Rad Laboratories, Hercules, CA) to extract total bacterial DNA. Aliquots of extracted DNA were subjected to polymerase chain reaction (PCR).
-
2.
Confirmation of bacterial DNA
First, to confirm that bacterial DNA was obtained appropriately, PCR amplification of the 16S ribosomal RNA gene (16S rDNA) was performed. The PCR mixture (25 μL) contained 12.5 μL of 2× AmpliTaq Gold® 360 Master Mix (Applied Biosystems, Carlsbad, CA), 10 pmol of forward and reverse universal primers (forward 5′-GTG STG CAY GGY TGT CGT CA-3′ and reverse 5′-ACG TCR TCC MCA CCT TCC TC-3′) [7], and a 2.5-μL aliquot of extracted DNA. PCR cycles were as follows: initial cycle at 95 °C for 10 min; 35 cycles at 95 °C for 1 min, 56 °C for 1 min, and 72 °C for 2 min; and a final extension at 72 °C for 5 min. Amplified products were subjected to 2 % agarose electrophoresis, and 120-bp DNA fragments were confirmed by ultraviolet light after ethidium bromide staining.
-
3.
mecA detection by PCR
mecA detection by PCR was performed as described previously [4]. The PCR mixture (25 μL) contained 12.5 μL of 2× AmpliTaq Gold® 360 Master Mix (Applied Biosystems), 10 pmol of primers (forward 5′-TGC TAT CCA CCC TCA AAC AGG-3′ and reverse 5′-AAC GTT GTA ACC ACC CCA AGA-3′), and a 2.5-μL aliquot of extracted DNA. PCR cycles were as follows: initial cycle at 95 °C for 10 min; 35 cycles at 95 °C for 30 s, 52 °C for 30 s, and 72 °C for 1 min; and a final extension at 72 °C for 7 min. Amplified products were subjected to 2 % agarose electrophoresis, and 284-bp DNA fragments were confirmed by ultraviolet light after ethidium bromide staining.
Statistical analysis
Differences in mecA carrier frequencies were compared by Fisher’s exact test or ANOVA using the statistical software IBM® SPSS® Statistics version 21 (IBM Corporation, NY). In all analyses, P < 0.05 was taken to indicate significance.
Results
Confirmation of bacterial DNA by PCR detection of 16S rDNA
The 16S rDNA PCR-amplified fragment was detected from 191 samples out of 209 prepared DNA samples from 59 HCT patients. The 16S rDNA was not detected in 18 samples, and mecA was also not detected from all these 18 samples in the following analysis. It was considered that in these 18 samples, the bacterial gene sample could not be collected appropriately because some technical error might have occurred in the sample correction due to the patients’ condition; therefore, these were excluded from further analysis. The collection rate of bacterial DNA from the oral mucosal swab was 91.4 %. The 16S rDNA fragment was successfully amplified by PCR from all samples from the control group (n = 52).
Frequencies of mecA carriers in groups of HCT patients and control subjects
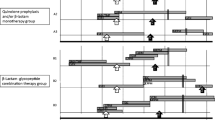
The frequency of mecA carriers in whom mecA was detected at least once during the study period (days −7 to +20 from HCT) was compared with that of the control group. The results are shown in Table 2 and Fig. 1. Seventy-six percent (45/59) of HCT patients carried mecA, while none of the control subjects had mecA. The difference in frequency of mecA carriers between HCT patients and control subjects was significant (P < 0.01 Fisher’s exact test).
Frequency of HCT patients in whom mecA was detected at least once during the HCT period, and transition of the frequency of mecA carriers in HCT patients. The difference in frequency of mecA carriers between HCT patients and control subjects was significant (# P < 0.01, Fisher’s exact test; the dagger represents the frequency of mecA carriers in whom mecA was detected at least once during days −7 to +20 from HCT). The detection frequencies of mecA increased significantly with time after HCT. The frequency of mecA carriers was 19.2 % at days −7 to −1 from HCT, while it increased significantly at days +7 to +13 and days +14 to +20, with frequencies of 60.9 and 63.2 %, respectively (*P < 0.01, ANOVA)
Transition of the frequency of mecA carriers in HCT patients
The transition of the frequency of mecA carriers on the oral mucosa before and after HCT is shown in Fig. 1. The detection frequencies of mecA increased significantly with time after HCT. The frequency of mecA carriers was 19.2 % on days −7 to −1 from HCT, while it was significantly increased from days +7 to +13 and days +14 to +20, with frequencies of 60.9 and 63.2 %, respectively (P < 0.01, ANOVA) (Table 3).
Discussion
The results of the present study indicated the presence of mecA in the oral cavity after HCT. The detection frequencies of mecA increased significantly with time after HCT. These results support those of our recent study indicating the detection of many CoNS and S. aureus with penicillin and beta-lactam resistance in the oral cavity after HCT [6].
In our recent study on antibiotic sensitivity of bacteria on the oral mucosa after HCT, CoNS with high degrees of resistance to penicillins and beta-lactams and methicillin-resistant S. aureus (MRSA) were detected [6]. We expected mecA detection based on our recent study using the culture method, while the frequency of mecA carriers on the oral mucosa was very high, over 60 % from days +7 to +20, which was beyond our expectation. This could be because a fourth-generation cephalosporin was mainly administered intravenously as empirical antibiotic therapy. A more in-depth analysis of these patients compared to HCT patients who were negative before HCT as well as the patients who got positive during treatment might be interesting and might corroborate our assumption that the administration of a fourth-generation cephalosporin was responsible for this increase in mecA detection. We will try to perform a multicenter study to increase subject number and would like to confirm our assumption.
The mecA gene complex is located on a unique molecular vector called the staphylococcal chromosome cassette (SCCmec) [3]. SCCmecs are considered to be transferred into S. aureus from a coagulase-negative species [6, 14]. The tendencies of mecA detection frequency may differ between institutes because of their policies of antibiotic use, while we speculate that the oral cavity just before and after HCT may be a reservoir and could be a transfer space of the genes regulating antibiotic resistance as mecA. Recent research strongly suggests that oral hygiene may also be a reasonable strategy to control methicillin-resistant CoNS to eventually lower the MRSA burden in medical facilities [8]. Maintenance of good oral hygiene after HCT may contribute to reducing the presence of genes regulating antibiotic resistance in the oral cavity and antibiotic-resistant bacterial infections.
In conclusion, mecA, which mediates penicillin and beta-lactam resistance, was detected from the oral mucosa immediately before and after HCT. The high detection frequency of staphylococci with resistance to penicillins and beta-lactams in our recent report was supported at the molecular level.
References
Bellm LA, Epstein JB, Rose-Ped A, Martin P, Fuchs HJ (2000) Patient reports of complications of bone marrow transplantation. Support Care Cancer 8:33–39
Chi F, Nolte O, Bergmann C, Ip M, Hakenbeck R (2007) Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int J Med Microbiol 297:503–512. doi:10.1016/j.ijmm.2007.02.009
de Lencastre H, Oliveira D, Tomasz A (2007) Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr Opin Microbiol 10:428–435. doi:10.1016/j.mib.2007.08.003
Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T (1992) Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett 298:133–136
Hughes WT, Armstrong D, Bodey GP, Bow EJ, Brown AE, Calandra T, Feld R, Pizzo PA, Rolston KV, Shenep JL, Young LS (2002) 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin Infect Dis 34:730–751. doi:10.1086/339215
Katayama Y, Takeuchi F, Ito T, Ma XX, Ui-Mizutani Y, Kobayashi I, Hiramatsu K (2003) Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J Bacteriol 185:2711–2722
Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, Arai H, Tanimoto I, Nishimura F, Takashiba S (2003) Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol 39:81–86
Schoenfelder SM, Lange C, Eckart M, Hennig S, Kozytska S, Ziebuhr W (2010) Success through diversity—how Staphylococcus epidermidis establishes as a nosocomial pathogen. Int J Med Microbiol 300:380–386. doi:10.1016/j.ijmm.2010.04.011
Soga Y, Maeda Y, Ishimaru F, Tanimoto M, Maeda H, Nishimura F, Takashiba S (2011) Bacterial substitution of coagulase-negative staphylococci for streptococci on the oral mucosa after hematopoietic cell transplantation. Support Care Cancer 19:995–1000. doi:10.1007/s00520-010-0923-9
Soga Y, Maeda Y, Tanimoto M, Ebinuma T, Maeda H, Takashiba S (2013) Antibiotic sensitivity of bacteria on the oral mucosa after hematopoietic cell transplantation. Support Care Cancer 21:367–368. doi:10.1007/s00520-012-1602-9
Soga Y, Saito T, Nishimura F, Ishimaru F, Mineshiba J, Mineshiba F, Takaya H, Sato H, Kudo C, Kokeguchi S, Fujii N, Tanimoto M, Takashiba S (2008) Appearance of multidrug-resistant opportunistic bacteria on the gingiva during leukemia treatment. J Periodontol 79:181–186. doi:10.1902/jop.2008.070205
Sonis ST (2004) The pathobiology of mucositis. Nat Rev Cancer 4:277–284. doi:10.1038/nrc1318
Sonis ST, Oster G, Fuchs H, Bellm L, Bradford WZ, Edelsberg J, Hayden V, Eilers J, Epstein JB, LeVeque FG, Miller C, Peterson DE, Schubert MM, Spijkervet FK, Horowitz M (2001) Oral mucositis and the clinical and economic outcomes of hematopoietic stem-cell transplantation. J Clin Oncol 19:2201–2205
Wielders CL, Vriens MR, Brisse S, de Graaf-Miltenburg LA, Troelstra A, Fleer A, Schmitz FJ, Verhoef J, Fluit AC (2001) In-vivo transfer of mecA DNA to Staphylococcus aureus [corrected]. Lancet 357:1674–1675
Acknowledgments
This study was supported by a Grant-in-Aid for Young Scientists (B no. 22791836 and no. 24792024 to YS) from the Japan Society for the Promotion of Science, and a Health and Labour Sciences Research Grant (24120701) from the Ministry of Health, Labour and Welfare, Japan
Conflict of interest
We have no conflicts of interest in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was partly presented as a poster at the MASCC/ISOO 2012 International Symposium on Supportive Care in Cancer in New York City on June 28–30, 2012.
Rights and permissions
About this article
Cite this article
Ebinuma, T., Soga, Y., Sato, T. et al. Distribution of oral mucosal bacteria with mecA in patients undergoing hematopoietic cell transplantation. Support Care Cancer 22, 1679–1683 (2014). https://doi.org/10.1007/s00520-014-2151-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00520-014-2151-1