Abstract
In this paper, we introduce an automatic and robust method to detect and identify Alzheimer’s disease (AD) using the magnetic resonance imaging (MRI) and positron emission tomography (PET) images. AD research as utilized with clinical and computer aid diagnostic tools has been strongly developed in recent decades. Several studies have resulted in many methods of early detection of AD, which benefit patient outcomes and new findings on the development of a deeper understanding of the mechanisms of this disease. Therefore, using the operation of electronic computers to diagnose automatically the incident of AD has served a vital role in supporting clinicians as well as easing significant elaboration on manual and subjectively AD diagnosing of clinicians for the patient’s beneficial outcomes. To this end, we propose a deep learning approach-based model of AD detection applying to MRI and PET images. Individually, we extract non-white matter of brain PET images, which are guided by MRI images as an anatomical mask. Before running the classification module, we build an unsupervised network entitled the high-level layer concatenation autoencoder to pre-train the network with inputs as three-dimensional patches extracted from pre-processed scans. The learned parameters are reused for a well-known convolutional neural network to boost up the training procedure. We conduct experiments on a public data set ADNI and classified a subject into one of three groups: normal control, mild cognitive impairment, and AD. Our proposed method outperforms for AD detection problem than other methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Alzheimer’s disease (AD) is an irreversible and progressive brain disorder that slowly destroys memory and thinking skills and eventually compromises the patient’s ability to carry out the simplest tasks. It is the most common form of dementia (Alzheimer’s Association 2014). The word dementia describes a set of symptoms that can include memory loss and difficulties with thinking, problem-solving, or language skills. These symptoms occur when the brain is damaged by certain diseases, including AD. Recently, there have been several tools and methods to diagnose the AD, such as conducting tests of memory, carrying out standard medical tests, and performing brain scans. The use of computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) is usually used for the brain scan system to rule out other possible causes for symptoms. The use of a structural MRI is a powerful tool because of high resolution and its ability to capture anatomical details of the brain, while the use of a PET is a promising indicator with discovering and investigating new radioactive tracers (Jack et al. 2001; McKhann et al. 2011). To take advantage of these images, the system first extracts the dominant features from brain images using feature extraction methods, and then a classifier is trained to identify which set of categories that an observation belongs. In general, a patient after classification is divided into three groups: AD, mild cognitive impairment (MCI), and normal control (NC).
In AD diagnosis, research in the use of a machine learning(ML) area was used to predict the AD using bio-masker extraction from multi-view of MRI, PET, and CT data (Bron et al. 2015). In the studies, the support vector machine (SVM) is the most popular ML method for classification applying after feature extraction process (Batmanghelich et al. 2009; Yang et al. 2011; Janousova et al. 2012; Rueda et al. 2012; Eskildsen et al. 2014). As dementia progresses, it is note that the volume of grey matter gradually decreases. This is called brain atrophy. For PET images in a normal ageing person, amyloid responses set on only white matter regions. In dementia, the amyloid uptake associates with a high magnitude that binds on grey matter. As dementia progresses, the intake of the amyloid in grey matter is greater. Therefore, the amyloid accumulation is normal in white matter only, but when the amyloid accumulation is also seen in grey matter, it is diagnosed as dementia. However, in reality, there are ambiguous cases when the amyloid uptake is beyond white matter if it is a normally ageing person, while it is not too high uptake in grey matter to diagnosis as a dementia patient.
While previous studies focused on MRI (Bron et al. 2015; Eskildsen et al. 2014; Kloppel et al. 2008; Suk et al. 2015) as an essential technique for AD detection, recent cases of AD’s clinical diagnosis with PET were reported notable results (Camus et al. 2012; Selnesa et al. 2012; Saint-Aubert et al. 2014; Heurling et al. 2015). Clinical results in (Heurling et al. 2015) report that PET agents targeting amyloid deposition cannot distinguish true grey matter uptake from white matter uptake. PET’s functionality mechanism is also based on mechanism of AD via radioactive visualization of PET agent. Therefore, it can more effectively detect the disease status of a person than the way which is based on only anatomical changes in brain structure as in MRI. Particularly, in a very old person (older than age 75), it is difficult to classify if this person’s brain is in normal or diseased since the volume of brain structure is commonly decreased according to age (Camus et al. 2012). It reveals that PET is necessary, especially for early AD detection, when chances in anatomy are hard to realize.
Typically, clinical PET-based AD detection is based on visualization by clinician experts. Many studies have focused on automatic detection of AD with the aid of statistical testing conducted with the role of experts in evaluations. These statistical values can be standard uptake value (SUV), standard uptake value ratio (SUVR), or the volume of each region of interest (ROI) (Saint-Aubert et al. 2014). However, these results are sensitive with group features as normalization, sampled scales, the variation of shape and the size of subject brains within the group. According to Noble and Scarmeas (2013), the PET imaging has a critical role as a functional technique that may enable clinicians to observe quickly and exactly activities related to AD of human brain, via diffusion of radioactive substances, such as the Pittsburgh compound B (PiB) (Klunk et al. 2004) or 18-FluoroDeoxyGlucose (FDG) (Mosconi et al. 2010). As stated in Mosconi et al. (2010), the FDG-PET images have been used to measure cerebral metabolic rates of glucose (CMRglc), a proxy for neuronal activity in AD. The CMRglc can be used to distinguish AD from other dementias, predict and track decline from normal cognition to AD, and identify individuals at risk for AD prior to the onset of cognitive symptoms.
Furthermore, because of the white matter degeneration, the amyloid uptake to white matter also can be changed according to the status of the disease (Selnesa et al. 2012). Therefore, it is useful to remove white matter (WM) from each PET image to avoid ambiguous amyloid uptake binding and consider the remaining regions as input to classify the status of AD. Based on these findings, we propose a fully automatic AD classification method using the discriminating features of non-WM PET images extracted by masking with MRI images from a same subject, which captured at the same time. Thanks to very quick response of PET agent, we can not only detect the status of diseased brain early, but also classify if a person’s brain is in normal or diseased accurately. By using the non-WM tissue, we solve the problem of the ambiguous amyloid based on MRI guide. In addition, we conduct a pre-trained strategy with a variant of convolutional autoencoder network, namely high-level layer concatenation autoencoder (HiLCAE) to learn hyper-parameters and apply them as an initialization for the following classifier network rather than using random initialization. Yosinski et al. (2017) proved that transferring the features even from distant tasks outperforms the random weights, because the transfer learning provides an effective way in training a large network using scarce training data without over-fitting. We use a well-known CNN architecture called VGG16 (Simonyan and Zisserman 2014) as AD classification method in our study. We conduct experiments for the classification of normal control (NC), mild cognitive impairment (MCI), and AD on ADNI data set. The results with classification accuracy are presented comparing to other methods.
The rest of this paper is organized as follows. Section 2 reviews the current literature on AD diagnosis. The detail of our proposed method is described in Sect. 3. The experimental setting and results are presented in Sect. 4. Finally, in Sect. 5, we conclude our research.
2 Related works
Several pattern classifiers have been conducted for predicting the AD status based on MRI, resting state MRI, PET images, and cerebrospinal fluid (CFS). Among many approaches, it is noted that the SVM has been used extensively in this case. The common point of these SVM-based methods is to first extract features on units such as voxels, ROI, grey matter, and then to manipulate the SVM to classify extracted features. Kloppel et al. (2008) study used grey matter (GM) voxels as features and trained an SVM to discriminate between AD and NC subjects. Gerardin et al. (2009) study used SVMs with linear kernels for classification of grey matter signatures and benchmarked results against the performance achieved by expert radiologists, which surprisingly were less accurate than the algorithm.
A concatenation of features from multiple modalities including MRI, PET, biological and neurological data into a vector (Kohannim et al. 2010) used an SVM as classifier. The whitening technique such as the independent component analysis (ICA) is seen as a feature extractor (Yang et al. 2011) and is also an effective SVM-based method for AD classification. An approach that combines penalized regression and data resampling for feature extraction prior to the classification using SVMs with Gaussian kernels is described (Janousova et al. 2012).
Recently, deep learning methods have also been explored in Genomic and Medical Image Data Analysis (Yu et al 2017). Gupta et al. (2013) used 2D convolutional neural network (CNN) for a slice-wise feature extraction of MRI scans. A multimodal deep Boltzmann machine (BM) was used (Suk et al. 2014) to extract one feature from each selected patch of the MRI and PET scans and predicts AD with an ensemble of SVMs. Li et al (2015) developed a multitask deep learning for AD classification and Mini–Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale–cognitive subscale (ADAS-cog) scoring by multimodal fusion of MRI and PET features.
There were many fusion algorithms utilized to combine two images into one modality to enhance the quality. It is also a good starting point to extract features or edge detection in order to improve performance. Lee et al. (2014) used the measure of sum of modified Laplacian map to compare and generate 2-level map, and then median-filtered to reduce effects of isolated noises. Agarwal and Bedi (2015) presented a hybrid technique using curvelet and wavelet transform to combine the images obtained by computed tomography (CT) scan and MRI. The resulted images obtain more information and additional data from the fused images. Differing from the above methods, when two images are treated equally and have no distinction of roles, our matching method treats the PET-MRI as a primary–subsidiary relation. In other words, the MRI is used as a mask and applied to PET image to take pure grey matter regions of PET, which is used later as an input of convolution neural network.
Motivated from the advantages of utilizing the multimodal technique with MRI and PET, as well as self-learning ability of deep learning network as CNN, we proposed an efficient automatic AD diagnosis method. In our study, we consider the non-white matter tissue of PET scans as a pattern classification of AD rather than using the whole images to revoke the ambiguous amyloid uptake. The use of a pre-trained network is also applied to our method to improve the performance of AD detection. Further, while the most of the methods study on binary classifiers such as MCI/NC and AD/NC, except Rueda et al. (2012), Gupta et al. (2013), and Eskildsen et al. (2014), we conduct further identification of classifiers with three classes of AD/MCI/NC.
3 AD detection using HiLCAE and 3D-VGG16
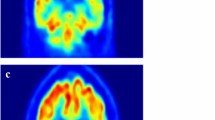
In this section, we introduce our proposed method to classify AD based on non-white matters using HiLCAE and VGG16. Figure 1 illustrates an ambiguous case between healthy brain and AD brain because amyloid uptake is beyond white matter. To solve this problem, preprocessing step is necessary to abolish white matter under brain tissue.
3.1 Preprocessing
We use the FDG-PET and MRI data downloaded from ADNIFootnote 1 data set with each pair of FDG-PET and MRI for same subject and captured at the same time. The MRI and PET images have undergone several pre-processed steps of research groups belonged to the ADNI. In detail, the MRI images are pre-processed by steps: gradwarp, B1 non-uniformity, and N3. Gradwarp means correction of image geometry distortion due to gradient model, and B1 non-uniformity is a correction procedure that uses B1 calibration scans to correct image intensity non-uniformity. Finally, a N3 histogram peak sharpening algorithm was applied to reduce intensity non-uniformity of images.
For the FDG-PET images, a procedure involving dynamic co-registering frames and acquiring averaging from baseline PET scan was conducted. PET images were reoriented as AC-PC correction into a standard \(160\times 160\times 96\) voxel image grid, having 1.5mm cubic voxels. These images underwent continued filtering with a scanner-specific filter function to procedure images of a uniform isotropic resolution of 8 mm full width at half maximum (FWHM).
The next steps are our own preprocessing steps as shown in Fig. 2, to acquire “non-white matter (WM) PET images”, that include:
-
1.
Co-register PET image to space of MRI image to result in same space and orientation for PET and MRI, as shown in Fig. 2a);
-
2.
Segment MRI to WM, grey matter probability maps by using segment module of SPM12Footnote 2 toolbox. The output of WM segmentation is shown in Fig. 2b);
-
3.
Extract WM from PET image by an order of sub-steps:
-
3.1
Inversely binarize WM map of MRI using threshold value. To choose a suitable threshold, we plot normalized histogram of all WM images and determine the average value where separates the dark (background) and bright (WM) regions. It is usually in the middle of two distributions, as shown in Fig. 3;
-
3.2
Cover full PET image by the mask in step 3.1, result in a “non-WM PET” image, as illustrated in Fig. 2c);
-
3.1
-
4.
Normalize “non-WM PET” images to a standard MNI template, using transformation matrix that calculated from normalizing coupled MRI to a MRI template in MNI space. This step is conducted by normalization module of SMP12 toolbox, and results are in all images with same size \(79\times 95\times 79\), i.e. 592, 895 voxels, since the original data scanned in various sizes of \(160\times 192\times 192\), \(166\times 256\times 256\), or \(164\times 256\times 256\), etc. The output after normalization is shown in Fig. 2d), which reduced an example of the co-registered size of \(166\times 256\times 256\) to the normalized size of \(79\times 95\times 79\). This work will save the computational time and memory cost, but without sacrificing the classification accuracy since the normalized image removes the non-informative region around brain and interpolated by 4th B-spline algorithm (Cheng et al. 2001).
3.2 HiLCAE
In this paper, we conduct a pre-trained network for a purpose of parameter initialization as shown in Huang et al. (2017). In practice, very few people train an entire deep network with random initialization, because it is relatively rare to have a data set of sufficient size. Instead, it is common to use a pre-trained network as an initialization or a fixed feature extractor for the task of interest. In this case, transfer learning provides an effective way in training a large network using scarce training data without over-fitting. Yosinski et al. (2017) proved that transferring the features even from distant tasks outperforms the random weights.
Figure 4 illustrates a schematic architecture of our proposed pre-trained network, called a high-level layer concatenation autoencoder (HiLCAE). This network is a variant of convolutional autoencoder (CAE) network. CAE of (Turchenko and Luczak 2017) is the architecture of choice for analysing structural data like images and 3D volumes. The convolution operator allows filtering an input signal in order to extract some part of its content. The autoencoders in their traditional formulation do not take this information into account. The fact is that a signal can be seen as a sum of other signals. The CAE, instead, uses the convolution operator to exploit this observation. They learn to encode the input in a set of simple signals and then try to reconstruct the input from them. The difference between our HiLCAE and the traditional CAE is that we concatenate the high-resolution feature from the encoding layer with the corresponding decoding layer. A successive convolution layer can then learn to assemble a more precise output based on this information. Moreover, the deconvolution part has also a large number of feature channels, which allow the network to propagate context information to higher-resolution layers.
In Fig. 4, HiLCAE composes two convolutional pooling layers for each encoder and decoder. Each layer includes two convolutional sub-layers with kernel size of \(3^3\) and activation function of rectified linear unit (ReLU) following by a max-pooling sub-layer to down-sample the output of convolutional sub-layer by \(2^3\) voxel. This procedure keeps only the highest value per \(2^3\) cude. After that, there are two minor layers entitled as the “flatten” and “bottleneck” layers. The “flatten” layer is used to spread 3D data onto the 1D vector, while the “bottleneck” layer is used for the nonlinear dimensionality reduction which usually contains few nodes compared to the previous layers. Next is the decoding layer, which has the same structure of convolutional operation to the encoding layer, but replaces the max-pooling sub-layer with the up-sampling sub-layer with size of \(2^3\) voxel. Besides, the network does not have any fully connected layers and only uses the valid part of each convolution. The cost function for backpropagation is mean spared error (MSE) as presented in (Nielsen 2015). Statistical gradient descent (SGD) by (Bottou 2010) is used to minimize the cost function. To reconstruct the image, we used the sigmoid formula as an activation function. After training, the learned weights and bias of encoding structure are used to initialize the next classification network in Sect. 3.3.
3.3 AD classification using adaptive 3D-VGG16
The visual geometry group (VGG) network (Simonyan and Zisserman 2014) is a convolutional neural network that won the ImageNet Competition in 2015 in the localization and classification categories. The VGG-16 is a 16-layer neural network and is usually used as pre-trained model for image recognition. However, to take advantage of the deep network, we built a 3D-VGG16 as classifier for identifying AD. We also adapted the convolutional layers and fully connected layers rather than using the original architecture of VGG-16 to avoid an over-fitting problem, which occurs when the amount of sample data is limited.
The extension of 2D-VGG16 to 3D introduces significant challenges: an increased number of parameters and important memory and computational requirements. Furthermore, 2D inputs accommodate using pre-trained nets, either directly or via transfer learning. However, an important drawback of such an approach is that anatomical context in directions orthogonal to the 2D plane is completely discarded. As discussed recently in Milletari et al. (2017), considering 3D data directly, instead of slice-by-slice, can improve the performance on working with neuroimaging.
Considering these facts, we proposed a microarchitecture of an adaptive 3D-VGG16 as described in Fig. 5, where the input is an 3D information of non-WM PET image with a size of \(79\times 95\times 79\). As shown in this figure, the 16 layers are divided into six subsets. The first two subsets include two convolutional layers with kernel size of \(3\times 3\times 3\), which are set up by the learned hyper-parameters (weights and bias) from the pre-trained HiLCAE, while the rest of the layers use random initialization, and a max-pooling layer to reduce the convolutional map by half. The next three subsets comprise three convolutional layers with the same kernel size and a \(2\times 2\times 2\) max-pooling layer. The last subset contains a fully connected layer and a softmax layer for the AD classification. In this paper, the output at every node is determined by a ReLU function. SGD is also applied to update the weights during training. Additionally, we execute four classifiers including binarized class and multi-class: AD/NC, AD/MCI, MCI/NC, and AD/MCI/MC. Therefore, the softmax layer in Fig. 5 can change from two nodes to three nodes in case of multi-class.
4 Experimental results
4.1 Materials and experimental setting
For the experiments, we used a publicly available ADNI data set on the web, as presented in Sect. 3.1. Specifically, we consider MRI and FDG-PET images acquired from 193 AD scans, 215 MCI scans, and 207 NC scans, where one subject can have several scan images. The total 615 scans are divided into training, validation, and test sets with 80%, 10%, and 10% of data, respectively. Detail of data set is described in Table 1.
All experiments were conducted five times, and we reported the average performance. We used the KerasFootnote 3 and TensorflowFootnote 4 framework for training the HiLCAE and fine-turn 3D-VGG16 networks. For the HiLCAE network, the number of convolutional pooling (conv-pool) set is changed from one to five corresponding to five conv-pool sets of VGG-16 network. The best performance is obtained when implementing with the first two conv-pool sets. Further, we trained each experiment during 5000 epochs on a GPU device and completed after 8\(\sim \)22 h, depending on the patch sizes. In our approach, the HiLCAE is expected to exploit latent presentations that may disentangle hidden factors of PET, controlling variability of images. We train the HiLCAE on a set of randomly selected 3D patches of size v extracted from non-white matter PET images. We experimented with many settings of v, and performance is reported as the lowest MSE for \(v = 25\), as shown in Fig. 6. We selected randomly 100 scans from each class (total scans are 300 for three classes) and extract 100 patches with size of 25-by-25-by-25 for each scan. Therefore, there are 30,000 patches in total, which divided into three sets: 24,000, 3000, and 3000 patches for training, validation, and test sets, respectively. For 3D-VGG16, we ran 500 epochs and configure an early stopping if there is no change of loss on validation set after 100 epochs. The learning rate is fixed by 1e-5 for both networks.
4.2 Performance
Table 2 reports the performance on four classifiers with different modalities on the test set. We found a result on the single and combined modalities of MRI and PET images. The results show that, for all classifiers, the non-WM PET model obtains better average accuracy (94.48%) than using only the whole brain of MRI or PET model (82.07 and 84.27%, respectively). It means that the proposed model significantly improves the accuracy of AD classification because of the noise reduction from WM. In addition, we compare the performance of our study with and without pre-trained network. Table 3 presents an amendment in the AD detection when using pre-trained model. Particularly, the accuracy of the four classifiers AD/NC, AD/MCI, MCI/NC, and AD/MCI/NC without pre-trained network is 87, 83.66, 87.52, and 81.63%, respectively. While the performance with the pre-trained CAE is 95.47% for AD/NC, 89.33% for AD/MCI, 91.05% for MCI/NC, and 88.72% for AD/MCI/NC. Specially, using the pre-trained HiLCAE, the accuracy increases to 98.8, 93, 95, and 91.13% respective to AD/NC, AD/MCI, MCI/NC, and AD/MCI/NC classifiers comparing to the result without using pre-trained model. The bold in Tables 2 and 3 indicate the average accuracy over the four classifiers.
Table 4 shows a comparison of the proposed method, the HiLCAE+3D-VGG16, and the state-of-the-art methods. The bold in this table shows the highest accuracy on each classifier: AD/MCI, MCI/NC, AD/NC, and AD/MCI/NC. The results also indicate that the HiLCAE+3D-VGG16 outperforms to other methods in terms of AD detection. Though using multimodality strategy causes time-consuming effort on the preprocessing step, the most relevant information from pure grey matter region of FDG-PET images helps finding discriminating features and remove outliers features of white matter which exist on single modality.
5 Conclusion
In this paper, we proposed an end-to-end AD classification system based on convolutional autoencoder and convolution neural networks using a combination of MRI and FDG-PET scans. By using the pre-trained weights from autoencoder network, we can boost up the classification process in the CNN. Our experiments indicate that the fusion approach has the potential to capture crucial local 3D amyloid uptake patterns without redundant, noisy information of white matter. Our proposed method also enhances correctness of using whole PET, also facilitating early detection AD rather than using MRI standalone, when anatomical changes occur slowly and are difficult to realize rather than functional changes occur inside a subject’s brain. For future works, we plan to build a deeper network for not only the HiLCAE but also the VGG16 to improve the classification performance. Further, we aim to develop a robust system that can deal with the whole-brain challenge.
Notes
Available at https://ida.loni.usc.edu. As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this paper.
References
Agarwal J, Bedi SS (2015) Implementation of hybrid image fusion technique for feature enhancement in medical diagnosis. Human-centric Comput Inf Sci 5:3
Alzheimer’s Association (2014) Alzheimer’s disease facts and figures. Alzheimer’s & Dementia, pp e47–e92
Batmanghelich N, Taskar B, Davatzikos C (2009) A general and unifying framework for feature construction, in image-based pattern classification. Inf Process Med Imaging 21:423–434
Bottou L (2010) Large-scale machine learning with stochastic gradient descent. In: COMPSTAT’2010, pp 177–186
Bron EE, Smits D, van der Flier WM et al (2015) Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: the CAD dementia challenge. NeuroImage 111:562
Camus V, Payoux P, Barr L et al (2012) Using pet with 18f-av-45 (florbetapir) to quantify brain amyloid load in a clinical environment. Eur J Nuclear Med Mol Imag 39:621–631
Cheng F, Wang X, Barsky BA (2001) Quadratic b-spline curve interpolation. Comput Math Appl 41:39–50
Eskildsen SF, Coupé P, Fonov V, Collins DL (2014) Detecting alzheimer’s disease by morphological MRI using hippocampal grading and cortical thickness. In: Proceedings of the 2014 MICCAI workshop challenge on computer-aided diagnosis of dementia based on structural MRI data, Boston, MA, pp 38–47
Gerardin E, Chtelat G, Chupin M et al (2009) Multidimensional classification of hippocampal shape features discriminates alzheimer’s disease and mild cognitive impairment from normal aging. NeuroImage 47:1476–1486
Gupta A, Ayhan M, Maida A (2013) Natural image bases to represent neuroimaging data. In: the 30th international conference on machine learning, pp 987–994
Heurling K, Buckley C, Vandenberghe R et al (2015) Separation of -amyloid binding and white matter uptake of 18F-flutemetamol using spectral analysis. Am J Nucl Med Mol Imaging 5(5):515–526
Huang Z, Pan Z, Lei B (2017) Transfer learning with deep convolutional neural network for sar target classification with limited labeled data. Remote Sens 9(9):907
Jack CR, Albert MS, Knopman DS et al (2001) Introduction to the recommendations from the national institute on aging-alzheimer’s association workgroups on diagnostic guidelines for alzheimer’s disease. Alzheimers Dement 7(3):257–262
Janousova E, Vounou M, Wolz R et al (2012) Biomarker discovery for sparse classification of brain images in alzheimer’s disease. Ann BMVA 2012:1–11
Kloppel S, Stonnington C, Chu C et al (2008) Automatic classification of MRI scans in alzheimer’s disease. Brain 131(3):681–689
Klunk WE, Engler H, Nordberg A et al (2004) Imaging brain amyloid in alzheimer’s disease with pittsburgh compoundb. Ann Neurol 55(3):306–319
Kohannim O, Hua X, Hibar DP et al (2010) Boosting power for clinical trials using classifiers based on multiple biomarkers. J Converg 31:1429–1442
Lee SH, Jung KH, Kang DW et al (2014) Pixel-based fusion algorithm for multi-focused image by comparison and filtering of sml map. Neurobiol Aging 5:28–31
Li F, Tran L, Thung KH et al (2015) A robust deep model for improved classification of ad/mci patients. IEEE J Biomed Health Inform 19:1610–1610
Lin S, Cai W, Pujol S, et al. (2014) Early diagnosis of alzheimer’s disease with deep learning. In: IEEE 11th international symposium on biomedical imaging, pp 1015–1018
McKhann GM, Knopman DS, Chertkow H et al (2011) The diagnosis of dementia due to alzheimers disease: recommendations from the national institute on aging-Alzheimers association workgroups on diagnostic guidelines for alzheimer’s disease. Alzheimers Dement 7(3):263–269
Milletari F, Ahmadi SA, Kroll C et al (2017) Hough-cnn: deep learning for segmentation of deep brain regions in mri and ultrasound. Comput Vis Image Underst 164:92–102
Mosconi L, Berti V, Glodzik L et al (2010) Pre-clinical detection of alzheimer’s disease using fdg-pet, with or without amyloid imaging. Alzheimers Dement 20(3):843–854
Nielsen M (2015) Using neural nets to recognize handwritten digits. Neural Networks and Deep Learning, chap 1
Noble JM, Scarmeas N (2013) Application of pet imaging to diagnosis of Alzheimer’s disease and mild cognitive impairment. Int Rev Neurobiol 84:133–149
Rueda A, Arevalo J, Cruz A, et al. (2012) Bag of features for automatic classification of alzheimer’s disease in magnetic resonance images. In: PPIACVA, pp 559–566
Saint-Aubert L, Nemmi F, Pran P et al (2014) Comparison between pet template-based method and mri-based method for cortical quantification of florbetapir (av-45) uptake in vivo. Eur J Nuclear Med Mol Imag 41:836–843
Selnesa P, Fjellc AM, Gjerstade L et al (2012) White matter imaging changes in subjective and mild cognitive impairment. Eur J Nuclear Med Mol Imag 41:112–121
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Suk HI, Lee SW, Shen D, ADNI, (2014) Hierarchical feature representation and multimodal fusion with deep learning for ad/mci diagnosis. NeuroImage 101:569–582
Suk HI, Lee SW, Shen D (2015) Latent feature representation with stacked auto-encoder for ad/mci diagnosis. Brain Struct Funct 220:841–859
Suk HI, Lee SW, Shen D (2016) Deep sparse multi-task learning for feature selection in alzheimer’s disease diagnosis. Brain Struct Funct 221(5):2569–2587
Turchenko V, Luczak A (2017) Creation of a deep convolutional autoencoder in caffe. In: 9th IEEE International conference on intelligent data acquisition and advanced computing systems: technology and applications (IDAACS), vol 2, pp 651–659
Yang W, Lui RML, Gao JH et al (2011) Independent component analysis-based classification of alzheimer’s disease MRI data. J AD 24(4):775–783
Yosinski J, Clone J, Bengio Y, et al. (2017) How transferable are features in deep neural networks? In: The 27th International conference on neural information processing systems, pp 3320–3328
Yu N, Yu Z, Gu F et al (2017) Deep learning in genomic and medical image data analysis: challenges and approaches. Inf Process Syst 13:204–214
Acknowledgements
This research was supported by the MSIP (Ministry of Science, ICT and Future Planning), Korea, under the ITRC (Information Technology Research Center) support programme (IITP-2017-2016-0-00314) supervised by the IITP (Institute for Information & communications Technology Promotion) and the Korean government (MSIP) (NRF-2017R1A2B4011409).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by G. Yi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vu, TD., Ho, NH., Yang, HJ. et al. Non-white matter tissue extraction and deep convolutional neural network for Alzheimer’s disease detection. Soft Comput 22, 6825–6833 (2018). https://doi.org/10.1007/s00500-018-3421-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-018-3421-5