Abstract
One of the key problems of computer-aided diagnosis is to segment specific anatomy structures in tomographic images as fast and accurately as possible, which is an important step toward identifying pathologically changed tissues. The segmentation accuracy has a significant impact on diseases diagnosis as well as the therapeutic efficacy. This paper presents a fast and robust weak-supervised pulmonary nodule segmentation method based on a modified self-adaptive FCM algorithm. To improve the traditional FCM, we firstly introduce an enhanced objective function, which computes the membership value according to both the grayscale similarity and spatial similarity between central pixels and neighbors. Then, a probability relation matrix between clusters and categories is constructed by using a small amount of prior knowledge learned from training samples. Based on this matrix, we realize a weak-supervised pulmonary nodules segmentation for unlabeled lung CT images. More specifically, the proposed method utilizes the relation matrix to calculate the category index of every pixel by Bayesian theory and PSOm algorithm. The quantitative experimental results on a test dataset, including 115 2-D clinical CT data, demonstrate the accuracy, efficiency and generality of the proposed weak-supervised strategy in pulmonary nodules segmentation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer, resulting from air pollution or smoking habits, has become one of the highest rates of incidences and mortality among all types of cancer. The statistic data provided by National Cancer Institute and Ministry of Health indicate that the mortality rate of lung cancer in China has increased 465% in the past 30 years. Unfortunately, this disease is often diagnosed at late stage due to the lack of symptoms and the poor prognostic. Therefore, computer-aided diagnosis (CAD) on lung cancer has been an increasingly important topic in the field of medical image processing.
At present, computed tomography (CT) is the most widely used technique to diagnose lung diseases among various medical imaging types (Puderbach and Kauczor 2008). As the early symptoms of lung cancer, pulmonary nodules have become an important research object. In the pulmonary nodules detection, the automatic extraction and assisted diagnosis by means of spiral CT play a significant role (Doi 2007). More importantly, it can improve the repeatability of diagnosis and consistency of image interpretation to some extent, and provide a quantitative analysis method for clinician, such as image segmentation and quantification of characteristics (Li et al. 2014). The requirement for obtaining a precise diagnosis of lung cancer has motivated many researchers to investigate automatic methods for pulmonary nodules detection and segmentation.
Recently, a growing numbers of pulmonary nodule segmentation methods have been proposed in the literature, which can be roughly categorized as six types: (1) Threshold methods (Reeves et al. 2006; Stelmo et al. 2012). For example, Tachibana and Kido (2006) proposed a threshold method for initial pulmonary nodule segmentation and then used the watershed algorithm for accurate segmentation. Xia et al. (2016) reveals that the histogram of the differences between pixel gray values is smoothed by the stego bits despite a large distance between the pixels. (2) Morphological methods (Dong and Peng 2014; Kubota et al. 2011; Gu and Sheng 2016). The work of Kishore and Satyanarayana (2013) used edge detection operator and morphological operations to segment lung nodules. (3) Variable model methods (McIntosh and Hamarneh 2012; Wen et al. 2015). For example, Dehmeshki et al. (2007) proposed an automatic detection method based on spherical shape of the structuring element sensitive detection. (4) Filter methods. For example, Li et al. (2008) proposed multi-scale dot filter, which can be used to enhance pulmonary nodules with the inhibition of angiogenesis. (5) Clustering methods (Murphy et al. 2009; Ortiz et al. 2014). For example, Zhang et al. (2012) proposed an improved FCM algorithm based on the histogram of the given image, which includes two phases. However, most of these methods perform well only for solitary pulmonary nodules (SPN) in the course of pulmonary nodule segmentation. In clinic, except for SPN, pulmonary nodules could also occur in forms of pleural adhesion, vascular adhesion, ground-glass opacity (GGO) and cavity type, and these types of nodule have higher probability of malignancy than SPN. In fact, it is a difficult to realize fast, correct and automatic segmentation of the nodules due to there being no distinguishing features between pulmonary nodules and surrounding tissues.
The well-known FCM is a classic and widely used segmentation algorithm, which applies fuzziness to the membership judgment of pixels and is consistent with human perception and convenient for realization. But the conventional FCM performs poor in low-contrast, in-homogeneity and noisy images as it does not take into account any spatial information and neighborhood correlation (Balafar et al. 2010; Chuang et al. 2006; Shi et al. 2009). In order to overcome these problems, many researchers have introduced spatial information through modifying the objective function or altering the distance form measurement between pixels and cluster centers. Ahmed et al. (2002) modified FCM objective function by incorporating spatial constraints (FCM_S). However, FCM_S lacks enough robustness to noise and outliers and is not suitable for revealing non-Euclidean structure of the input data due to the use of Euclidean distance. Two improved variants, FCM_S1 and FCM_S2, were proposed with an intent to simplify the computation of parameters (Chen and Zhang 2004), which were further extended to the corresponding kernelized versions. An enhanced FCM (EnFCM) algorithm proposed by Szilagyi et al. (2003) speeds up the clustering process on the basis of the gray-level histogram instead of pixels, which uses a linearly weighted sum image formed from both original image and each pixel’s local neighborhood average gray level. Hence, the computational time of EnFCM can be reduced greatly since the number of gray levels in an image is much smaller than that of pixels. A fast generalized FCM (FGFCM) algorithm (Cai et al. 2007) introduces the spatial information combining the intensity of the local pixel neighborhood and the number of gray levels in an image, so that the quality of segmentation result is well enhanced with a low computational cost. Krinidis and Chatzis (2010) proposed a novel robust fuzzy local information C-means clustering (FLICM) algorithm by incorporating the local spatial- and gray-level information, which is free of the empirically adjusted parameters and enhances the clustering performance. Recently, by estimating and correcting the heterogeneity of the magnetic field image (bias) to minimize the noise effects, Bakhshali (2016) presented an improved FCM method based on information theoretic clustering.
Although these modified FCM algorithms have improved the performance of the conventional FCM, there are still the following disadvantages: (1) The desired convergence needs a large number of iterations; (2) Different types of pulmonary nodules with complicated features lead to the worst segmentation performance, including low-contrast nodules, vasculature attachment and close vicinity nodules. So, we proposed a modified self-adaptive FCM algorithm that defines a new objective function (Liu et al. 2015). It computes the membership value by combining the grayscale similarity and spatial similarity between the central pixels and neighborhood, in which the local grayscale similarity depending on the difference between the gray scale of central pixels and neighborhood is used to suppress the noise and speed convergence, and the local spatial similarity depending on the size of neighborhood, as well as the difference between the average gray scale of neighborhood and central pixels, is adopted to enhance the adaptivity of algorithm. These modifications effectively improve the convergence rate and the adaptivity to spatial characteristics of pulmonary nodule edges.
However, the modified self-adaptive FCM algorithm mentioned above is a kind of unsupervised learning method. In fact, it is possible to further improve its performance by using some prior segmentation information, especially for pleural adhesion, vascular adhesion and ground-glass opacity (GGO) nodules. Therefore, it is significant that only a small amount of labeled classification prior and constraint prior can instruct a fast and robust pulmonary nodules segmentation work. In this paper, we propose a weak-supervised pulmonary nodule segmentation method, which constructs a probability relation matrix between clusters and categories information according to training samples and calculates the category index of every pixel of testing samples by Bayesian theory and modified particle swarm optimizer (PSOm) algorithm (Shi and Eberhart 1998).
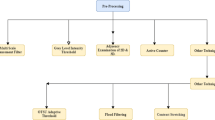
As illustrated in Fig. 1, the proposed method for segmenting the pulmonary nodule consists of two phases, i.e., the training stage and testing stage. First of all, according to the spatial information and grayscale information, the modified FCM function updates the fuzzy membership degree of center pixels during each iteration with a faster speed. In this way, a probability relation matrix can be constructed according to cluster centers and a small amount of manual labeled information. The next stage is the separation of the tissues resembling the pulmonary nodules based on the logical relationship between clusters and categories, which is a weakly-supervised segmentation for unlabeled CT images.
The rest of this paper is organized as follows. Section 2 briefly introduces a modified self-adaptive FCM algorithm followed by some experimental results and discussions. Section 3 presents the proposed fast weakly-supervised learning model, which is essentially a two-phase classification algorithm by using a small amount of manual information. Section 4 reports the experimental results, followed by some discussions, on four types of pulmonary nodules in terms of precision, computational speed and sensitivity to free parameters. Finally, Sect. 5 concludes this paper with the future work.
2 Self-adaptive FCM algorithm
In the chest CT images, we should extract the lung tissues from other organs according to their gray values, such as skeletons, mediastina and trachea. In general, it is different to distinguish pulmonary nodules from blood vessels (vascular adhesion) and bronchi, which have similar density information. From the visual information, the tomography image region of pulmonary nodules is bright in center and dark in peripheral area, where the bright pixels reflect the high-density character of nodules, conversely for the low-density areas.
In order to effectively distinguish the density differences between nodules and structures that resembling nodules, and remove the weak edge distribution of peripheral tissues, we modify the traditional FCM algorithm by integrating the membership value from both the grayscale similarity and spatial similarity between the central pixels and neighborhood into the objective function of FCM. This algorithm can be divided into two steps. Firstly, a 2D vector, which represents the spatial relationship between a central pixel and its neighborhood, is constructed. Secondly, the spatial information and grayscale information of the neighborhood around the central pixel are used to determine an objective function.
As a typical clustering algorithm based on division, the traditional FCM algorithm aims to divide N pixels into C fuzzy groups for image I. It searches the cluster center of each group by updating the membership value of each pixel iteratively. This update is implemented by means of minimizing the objective function, which is defined as:
where C is the number of cluster, \(v_{k}\) represents the center of the \(k \mathrm{th}\) clusters, N is the number of pixels in image I, \(x_{i}\) denotes the \(i \mathrm{th}\) sample of I, \(\mu _{ki}\) is the membership value of the \(i \mathrm{th}\) sample with respect to the \(k \mathrm{th}\) cluster, which is defined as \([(\sum _{j=1}^{C}(\frac{|x_{i}-v_{k}|}{|x_{i}-v_{j}|}))^{m}]^{-1}\).
An intuitive interpretation of this objective function is to assign higher fuzzy membership to pixels whose gray scales are close to those of specific clustering centers, and assign lower fuzzy membership to pixels whose grayscale values are distinct from those centers.
But it is not suitable for complex pulmonary nodule segmentation. The reasons are as follows: (1) The grayscale characteristics of different tissues in lung CT image make it difficult to obtain a smooth and closed segmentation edge; and (2) the executive efficiency of the traditional FCM algorithm is low due to a number of iterations for convergence. Other different variants, such as FCM_S, FCM_S1, FCM_S2, FGFCM and EnFCM, take into account the information of neighborhood pixels that belong to the central pixels and introduce a control parameter for the intensity of neighborhood information. This parameter has an important influence on the performance of FCM. However, its value maintains same during the whole process of segmenting an image. Obviously, this selection strategy for the control parameter can not reflect the features of neighborhood pixels.
In fact, this parameter can be determined by the following factors: (1) The local grayscale similarity, i.e., the parameter should depend on the difference between the gray scale of central pixels and neighborhood, in order to suppress the noise and speed up the convergence; (2) the local spatial similarity, i.e., it should also depend on the size of neighborhood, as well as the difference between the average gray scale of central pixels and neighborhood, in order to enhance the adaptivity of algorithm. Based on the discussions above, the modified self-adaptive FCM algorithm can be described as Algorithm 1. The computational efficiency of this algorithm is significantly improved because the number of iterations is much less than the traditional FCM and FCM_S. Besides, both EnFCM and FGFCM algorithms transform the segmentation based on pixels into the segmentation based on grayscale levels. Due to the fact that the grayscale level (0-255) is much less than the number of pixels, they become much faster.

Then, we compared Algorithm 1 with other five related algorithms (FCM, FCM_S, EnFCM, FGFCM and FLICM) about further segmentation performance. In these algorithms, the number of clusters C is an essential variable. We empirically set \(C =4\), and it is effective to distinguish the different parts of lung parenchyma. Table 1 lists the other optimal experimental parameters of each method, respectively, which are determined by extensive experiments. Note that m is the fuzzy weight exponent, which is used to generalize fuzzy clustering objective functions (Bezdek 1976). In this paper, m plays a role of suppressing noise and smoothing membership functions. The empirical range of m given by Bezdek is \(1.1\le m \le 5\). Due to having a suitable physical interpretation, we set \(m = 2\).
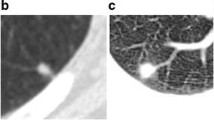
Figure 2 gives the segmentation results of isolated, vascular adhesion, pleural adhesion and ground-glass opacity pulmonary nodules by Algorithm 1. For each case, it provides nine comparison images, including the original CT image, its pulmonary parenchyma image, segmentation results of nodule from the manual labeling and six FCM algorithms. Note that the manual labeling is obtained by averaging the contour information labeled by four clinical radiologists, and it will be used as the ground truth in this part and Sect. 4. For example, Fig. 3 illustrates how to obtain the reference contour. We invite four radiologists to blindly read and annotate the location and character for each nodule by enlarging the image a number of times, for the purpose of providing manual segmentation. From multiple manual segmentation results, we construct the final single reference as ground truth that is included in at least \(50\%\) of the available segmentations.
From Fig. 2a, it can be found that, for an isolated pulmonary nodule, FCM and FGFCM are influenced by noise severely, and there still exists noise in the results. Although FCM_S and EnFCM perform better than the two aforementioned algorithms, the segmentation results of objects and tiny blood vessels are much less than the ground truth. Our method effectively reduces the negative influence of noise and successfully separates objectives from background. Figure 2b shows all information of a vascular adhesion pulmonary nodule. FCM, FCM_S and EnFCM yield messy results that differ greatly from the manual ground truth. The main reason is that they do not eliminate the surrounding small extraneous information and noise. Furthermore, according to their boundary of blood vessels, it can be seen that there are boundary leakage occurrences to some extent. Our method can overcome these problems and perform well in segmenting the vascular adhesion pulmonary nodules. Figure 2c displays the results of pleural adhesion pulmonary nodule. The boundary shape obtained by our method is the closest to the ground truth, and those of other methods reveal few wrong segmentations to a certain extent. Because the connection area between the nodule and lung wall is especially delicate, it is difficult to distinguish the extended area of nodule and background of pulmonary parenchyma obviously. In Fig. 2d, we can find a ground-glass opacity (GGO) pulmonary nodule, which is indicated by a red arrow. It can be observed that the traditional FCM hardly segments this pulmonary nodule, because this method can only extract the brightest region around the center. FCM_S, EnFCM and FGFCM perform less than FLICM on aspects of nodule’s boundary convergence and tiny blood vessels. In this case, the result obtained by our method is superior to others.
Table 2 lists the average error rate \(A(C_{m},C_{0})\) of segmentation results by different algorithms on dataset described in Table 3. \(A(C_{m},C_{0})\) is defined as
where \(C_{m}\) represents the reference segmentation result contained in LIDC dataset (Armato et al. 2011) or obtained manually, e.g., Fig. 3c, and \(C_{0}\) is obtained automatically by different algorithm, e.g., our segmentation result given in Fig. 3d.
Furthermore, we use mean and std. to quantify the statistical performance of these algorithms for segmenting four types of nodules, where mean shows the average error rate of each algorithm, and std. compares the algorithm stability given by standard deviation. Although this modified FCM does not arrive the best level, it performs well for isolated nodules and GGO types obviously.
3 Fast weak-supervised segmentation method based on Bayesian and PSOm
In the previous section, we introduced the modified FCM, which can improve the efficiency and robustness of clustering. However, in this method, cluster centers are randomly chosen at the initial stage. It is obvious that this kind of initialization will degrade the performance of unsupervised clustering when it is applied to unlabeled samples. To address this issue, we propose a fast weak-supervised segmentation method by using a small amount of prior labeled information. The proposed algorithm can estimate the intrinsic logical relationship between clusters and categories for unlabeled images based on Bayesian theory, and then, it utilize the modified particle swarm optimizer (PSOm) to obtain cluster centers embedded in the objective function of FCM and category index from new samples efficiently.
Thus, after achieving the clusters from input space by above modified self-adaptive FCM algorithm, we can design a classifier. In the process of classification, the posterior probability that pixel \(x_{i}\) with respect to each category of test samples will be estimated based on the cluster \(v_{k}\) generated by Algorithm 1, and the output category index of \(x_{i}\) is calculated by:
where \(\omega _{l}\) means the \(l\mathrm{th}\) category obtained by prior labeled information, \(p(\omega _{l}\mid x_{i})\) is obtained based on structure information of sample clusters:
where \(v_{k}\) means the \(k\mathrm{th}\) cluster obtained by Algorithm 1, \(p(v_{k}\mid x_{i})\) is the cluster posterior probability of pixel \(x_{i}\):
where \(\mathrm{dist}()\) means the Euclidean distance.
Since \(p(\omega _{l}\mid v_{k},x_{i})\) in Eq. (4) is irrelevant to \(x_{i}\), it can be simplified as \(p(\omega _{l}\mid v_{k})\), which means the category posterior probability of the cluster. In this work, \(p(\omega _{l}\mid v_{k})\) can be defined as follows:
where \(p(v_{k})\) means the ratio that the pixel samples belong to the \(k\mathrm{th}\) cluster relative to the total number, i.e., \(Num(x\in v_{k})/N\); \(p(\omega _{l},v_{k})\) means the ratio that the samples being in both the \(k\mathrm{th}\) cluster and the \(l\mathrm{th}\) category relative to the total number, i.e., \(\mathrm{Num}(x\in \omega _{l}\bigcap x\in v_{k})/N\).
Obviously, the greater the value \(p(\omega _{l}\mid v_{k})\)is, the more the samples simultaneously belong to the \(k\mathrm{th}\) cluster and the \(l\mathrm{th}\) category. Thus, when the category index of training samples has been known, \(p(\omega _{l}\mid v_{k})\) depends on the cluster algorithm that is related to the cluster centers.
Finally, the algorithm constructs a probability relationship matrix P with the size \(C\times L\), which is defined as:
Furthermore, in order to enhance the performance, this paper employs the modified PSOm proposed by Shi and Eberhart in 1998. In the process of accelerating the calculation of cluster centers, particle \(v_{i}=[v_{i_{1}},v_{i_{2}},\ldots ,v_{i_{K}}]\) corresponds to a vector that composed of all cluster centers in each sample, here \(K=4\). The best location that each particle \(v_{i}\) experiences is \(p_{i}=[p_{i_1},p_{i_2},\ldots ,p_{i_N}]\). The best particle in the group is denoted by the subscript g. Finally, each particle updates its velocity based on the experiences from itself and whole group:
where \(\alpha _{1}\) and \(\alpha _{2}\) are speeding factors, whose empirical value is usually between 1 and 4; \(r_{1}\) and \(r_{2}\) are random values located at [0,1]; \(\beta \) is an inertia factor, which is used to balance the global and local searching courses.
Above all, this fast weak-supervised segmentation algorithm can be summarized as follows:

4 Experimental results and analysis
In the experiments, we use two test datasets: the LIDC2 datasetFootnote 1 and the lung CT image database.Footnote 2 The concrete information about these two sets is described in Table 3. We randomly choose five images per individual case from the experimental CT set as the training set, which includes totally 65 images and 84 pulmonary nodules. A random database partition is performed with 35, 12, 26 and 10 nodules of each type for training, and the rest of the database for testing.
4.1 Segmentation result of pulmonary nodules
In this subsection, we use 165 CT images and 363 nodules in total to test our method. The experimental results reveal that the error rate of our method running on training data is lower than that on testing data. For the purpose of objective analysis, we only use the testing data to illustrate the visual quality. In part of visual comparison, the representative segmentation results of isolated, vascular adhesion, pleural adhesion and ground-glass opacity(GGO) pulmonary nodules are shown in Figs. 4 and 5, respectively. Obviously, our weak-supervised segmentation method performs better because of the light prior information.
Generally, the blood vessels have similar gray scale with pulmonary nodules, and GGO pulmonary nodules have low contrast to backgrounds. The grayscale values of the pixels between pleura and pulmonary nodules are changed smoothly, which means a low contrast to the background. The traditional FCM algorithm segments pulmonary nodules by clustering similar data points in the characteristic space and does not take into account the interactions between neighboring pixels. It yields to a worse performance for complicated features. By introducing neighborhood information to update membership values, other four FCM algorithms can reduce the influence of noise to some extent. However, as considering spatial similarity and grayscale similarity, these algorithms fall into the local extremum and perform sensitive to the initial values.
4.2 Execution performance of algorithms
In order to further demonstrate the efficiency of the proposed method, Table 4−6 and Fig. 6 offer several statistical results on training data and testing data. Specifically, Table 4 lists the average number of iterations (the maximum number of iteration \(L_{\max }=100\) or threshold \(\varepsilon \) reaches the value in Table 4), and the corresponding running time of each algorithm.
Obviously, FGFCM and FLICM need more iterations to satisfy the convergence threshold \(\varepsilon \). As a result, the running time of FGFCM and FLICM is also longer than others. Other two algorithms FCM and FCM_S can converge with less iterations and higher efficiency. EnFCM algorithm can speed up the cluster processing, because it translates the original image into the average local neighborhood image by calculating the sum of linear weighted from its eight neighbor pixels. Since the grayscale levels of the pixels are typically 8-bit resolution (256 levels), this method can reduce the encoding length and shorten the time complexity significantly. However, it can be observed that the segmentation effect of EnFCM is not satisfied from Figs. 4 and 5. By comparison, our method needs the least iteration number and the shortest running time, which is consistent with the discussion described in Sect. 3, because of more accurate initial cluster centers during the iteration by learning.
Table 5 gives the average error rates \(A(C_{m},C_{0})\) of the five typical algorithms and our method, which are achieved by applying different algorithms to the CT image sets explained in Table 2. The mean and standard deviation of \(A(C_{m},C_{0})\) of our method are 0.18 and 0.17, respectively. It means that the error rate of our method is less than other algorithms. From Table 5, we can easily find that the performance has been greatly improved by introducing the weak-supervised learning, especially for the vascular adhesion and pleural adhesion nodules.
Table 6 further shows the comparison results about segmentation performance and efficiency with various reference methods. From top to bottom, the methods of Tachibana and Kido (2006), Kishore and Satyanarayana (2013), Dehmeshki et al. (2007), Li et al. (2008) are given, all of them are classical pulmonary nodule segmentation methods. Tachibana’s method is punished by the poor accuracy and low efficiency, because there is the process of complex space transformation and dynamic programming. Kishore’s method performs more accurate for vascular adhesion nodules, because the ROI lung area blood vessels and nodules from the major lung portion are extracted using different edge detection filters. Dehmeshki’s method is a typical template matching method based on genetic algorithm, but the low convergence speed of genetic algorithm is derived by a large of individual calculation so that it has lower accurate and operation speed. The result reported in (Li et al. 2008) is based on the selective nodule enhancement filter, which significantly enhances the difference of nodules and normal anatomic structures. Our method obtains the best overall performance by training process for a part of labeled information.
In addition, Fig. 6 demonstrates the boundary accuracy comparison between our method \(e_{a}\) and ground truth, where the test samples, corresponding to above four types pulmonary nodules (a)–(d), are 162, 97, 58 and 46, respectively. The horizontal ordinate represents the rate of wrong segmentation \((e_{m}-e_{a})/e_{m}\) (%), and the vertical ordinate represents the corresponding number of pulmonary nodules. A positive rate of wrong segmentation means under-segmentation resulting from the algorithm with respect to ground truth, and a negative rate of wrong segmentation means over-segmentation results.
5 Conclusion and future work
In this paper, a fast and robust weak-supervised pulmonary nodule segmentation method is proposed, for the purpose of ideal segmentation of vascular adhesion, pleural adhesion and GGO pulmonary nodules. It combines the modified FCM cluster algorithm and small amount of labeled category information. Specially, the proposed method improves the traditional FCM algorithm according to the boundary features. This modified self-adaptive FCM updates the membership value of central pixels based on the grayscale similarity and spatial similarity of neighborhood pixels and obtains all cluster centers quickly. Obviously, this method can solve some segmentation problems, for example, the segmentation performance reliance on the contrast between the objectives and background, the complexity on the weak edge of pulmonary nodules and adhesive tissues, etc. Secondly, according to the fact that the number of unlabeled sample is much larger than the labeled, our method analyzes the intrinsic logical relationships between the clusters and categories of training samples and realizes the weak-supervision of prior knowledge for classifying the unlabeled testing samples.
The proposed method can obtain more accurate initial cluster centers by learning, which is better than typical unsupervised FCM. Experimental results demonstrate that it can segment isolated, vascular adhesion, pleural adhesion and GGO pulmonary nodules fast, and perform better than traditional FCM, FCM_S, EnFCM, FGFCM and FLICM in segmentation results, computational efficiencies and error rates.
However, there are still some disadvantages about the proposed method. For example, it does not work well for segmenting the tiny nodules (\(diameter<1\,cm\)), and the results of GGO nodules and tightly adhered nodules are unstable. In the future works, we will try to solve these problems by extracting more characteristics of pulmonary nodules or referring to the deep learning strategy and also analyze the clinical symptoms of benign and malignant nodules comprehensively to provide real aided diagnosis for early detection of lung cancers. Besides, we will also extend the low-rank property (Guo et al. 2016) of images to segment the pulmonary nodules. With the development of big data and cloud computing, some data encryption algorithms (Li et al. 2015, 2014), fuzzy keyword search algorithm (Li et al. 2010) and other algorithms (Zhu et al. 2016) determine the image base of unknown format files and secure de-duplication algorithm (Li et al. 2014, 2015; Yan et al. 2016) have been proposed; we can obtain a larger and more accurate and non-redundant CT image set to help us improve the auxiliary diagnostic effect.
Notes
Provided by National Institute of Health’s Lung Imaging Database Consortium (LIDC), which is available at https://public.cancerimagingarchive.net/ncia/login.jsf
Provided by Shandong Provincial Qianfoshan Hospital
References
Ahmed MN, Yamany SM, Mohamed N, Farag AA, Moriarty T (2002) A modified fuzzy C-means algorithm for bias field estimation and segmentation of MRI data. IEEE Trans Med Imaging 21(3):193–199
Armato SG, McLennan G, Bidaut L et al (2011) The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931
Bakhshali MA (2016) Segmentation and enhancement of brain MR images using fuzzy clustering based on information theory. Soft Comput. doi:10.1007/s00500-016-2210-2
Balafar MA, Ramli AR, Mashohor S, Farzan A (2010) Compare different spatial based fuzzy c-mean (fcm) extensions for MRI image segmentation. In: The 2nd international conference on computer and automation engineering, pp 609–611
Bezdek JC (1976) A physical interpretation of fuzzy ISODATA. IEEE Trans Syst Man Cybern 6(5):387–389
Cai W, Chen S, Zhang D (2007) Fast and robust fuzzy C-means clustering algorithms incorporating local information for image segmentation. Pattern Recognit 40(3):825–838
Chen S, Zhang D (2004) Robust image segmentation using FCM with spatial constraints based on new kernel-induced distance measure. IEEE Trans Syst Man Cybern 34(4):1907–1916
Chuang KS, Tzeng HL, Chen S, Wu J, Chen TJ (2006) Fuzzy C-means clustering with spatial information for image segmentation. Comput Med Imaging Gr 30(1):9–15
Dehmeshki J, Ye X, Lin XY, Valdivieso M, Amin H (2007) Automated detection of lung nodules in CT images using shape-based genetic algorithm. Comput Med Imaging Gr 31(6):408–417
Doi K (2007) Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Gr 31(4–5):198–211
Dong F, Peng J (2014) Brain MR image segmentation based on local Gaussian mixture model and nonlocal spatial regularization. J Vis Commun Image Repress 25(5):827–839
Gu B, Sheng VS (2016) A robust regularization path algorithm for v-support vector classification. IEEE Trans Neural Netw Learn Syst. doi:10.1109/TNNLS.2016.2527796
Guo Q, Zhang C, Zhang Y, Liu H (2016) An efficient svd-based method for image denoising. IEEE Trans Circuits Syst Video Technol 26(5):868–880
Kishore VV, Satyanarayana RVS (2013) Performance evaluation of edge detectors - morphology based ROI segmentation and nodule detection from DICOM lung images in the noisy environment. In: IEEE 3rd international advance computing conference, pp 1131–1137
Krinidis S, Chatzis V (2010) A robust fuzzy local information C-means clustering algorithm. IEEE Trans Image Process 19(5):1328–1337
Kubota T, Jerebko AK, Dewan M, Salganicoff M, Krishnan A (2011) Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med Image Anal 15(1):133–154
Li J, Chen X, Li M, Li J, Lee PPC, Lou W (2014) Secure deduplication with efficient and reliable convergent key management. IEEE Trans Parallel Distrib Syst 25(6):1615–1625
Li J, Dang J, Wang Y (2014) Medical image segmentation algorithm based on quantum clonal evolution and two-dimensional tsallis entropy. J Comput Aided Des Comput Gr 26(3):465–471 (In Chinese)
Li J, Huang X, Li J, Chen X, Xiang Y (2014) Securely outsourcing attribute-based encryption with checkability. IEEE Trans Parallel Distrib Syst 25(8):2201–2210
Li J, Wang Q, Wang C, Cao N, Ren K, Lou W (2010) Fuzzy Keyword Search over Encrypted Data in Cloud Computing. In: Proceeding of the 29th IEEE international conference on computer communications, pp 441–445
Li J, Li J, Chen X, Jia C, Lou W (2015) Identity-based encryption with outsourced revocation in cloud computing. IEEE Trans Comput 64(2):425–437
Li Q, Li F, Doi K (2008) Computerized detection of lung nodules in thin-section CT images by use of selective enhancement filters and an automated rule-Based classifier. Acad Radiol 15(2):165–175
Liu H, Zhang C-M, Su Z-Y, Wang K, Deng K (2015) Research on a pulmonary nodule segmentation method combining fast self-adaptive FCM and classification. Comput Math Methods Med 2015:185726. doi:10.1155/2015/185726
McIntosh C, Hamarneh G (2012) Medial-based deformable models in nonconvex shape-spaces for medical image segmentation using Genetic Algorithms. IEEE Trans Med Imaging 31(1):33–50
Murphy K, Ginneken B, Schilham AMR, Hoop BJ, Gietema HA, Prokop M (2009) A large scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbor classification. Med Image Anal 13(5):757–770
Ortiz A, Gorriz JM, Ramirez J, Salas-Gonzalez D (2014) Improving MR brain image segmentation using self-organising maps and entropy-gradient clustering. Inf Sci 262:117–136
Puderbach M, Kauczor H (2008) Can lung MR replace lung CT. Pediatr Radiol 38(3S):439–451
Reeves AP, Chan AB, Yankelevitz DF (2006) On measuring the change in size of pulmonary nodules. IEEE Trans Med Imaging 25(4):435–450
Shi Y, Eberhart R (1998) A modified particle swarm optimizer. Evolut Comput Proc. doi:10.1109/ICEC.1998.699146
Shi Z, Zhou P, He L, Nakamura T, Yao Q, Itoh H (2009) Lung segmentation in chest radiographs by means of gaussian kernel-Based FCM with spatial constraints. In: The 6th international conference on fuzzy systems and knowledge discovery , pp 428–432
Stelmo M, Aristofanes CS, Rodolfo AN, Marcelo G (2012) Automatic segmentation of lung nodules with growing neural gas and support vector machine. Comput Biol Med 42(11):1110–1121
Szilagyi L, Benyo Z, Szilagyii S (2003) MR brain image segmentation using an enhanced fuzzy C-means algorithm. Int Conf Eng Med Biol Soc. doi:10.1109/IEMBS.2003.1279866
Tachibana R, Kido S (2006) Automatic segmentation of pulmonary nodules on CT images by use of NCI lung image database consortium. Medical Imaging 2006: Image Processing, 61440M, doi:10.1117/12.653366
Wen X, Shao L, Xue Y, Fang W (2015) A rapid learning algorithm for vehicle classification. Inf Sci 295:395–406
Xia Z, Wang X, Sun X, Liu Q, Xiong N (2016) Steganalysis of LSB matching using differences between nonadjacent pixels. Multimed Tools Appl 75(4):1947–1962
Yan F, Tan Y, Zhang QX, Wu F, Cheng Z, Zheng J (2016) An effective RAID data layout for object-based de-duplication backup system. Chin J Electron 25(5):832–840 (In Chinese)
Zhang XF, Zhang CM, Tang WJ (2012) Medical image segmentation using improved FCM. Sci China F 55(5):1052–1061
Zhang Y, Li G, Sun X, Zhou X (2009) Geometric active contours without re-initialization for image segmentation. Pattern Recognit 42(9):1970–1976
Zhu R, Tan Y, Zhang Q, Wu F, Zheng J, Xue Y (2016) Determining image base of firmware files for ARM devices. IEICE Trans Inf Syst E 99D(2):351–359
Acknowledgements
This work is supported by National Natural Science Foundation (61572286 and 61472220), Natural Science Foundation of Shandong Province (2016ZRB01143), University Independent Innovation Plan (201401216), Science and Technology Development Plan of Shandong province (2014GGX101037), and also supported by Fostering Project of Dominant Discipline and Talent Team of Shandong Province Higher Education Institutions. The authors also gratefully acknowledge the helpful comments and suggestions of the reviewers, which have improved the presentation significantly.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Hui Liu declares that she has no conflict of interest. Fenghuan Geng declares that he has no conflict of interest. Qiang Guo declares that he has no conflict of interest. Caiqing Zhang declares that he has no conflict of interest. Caiming Zhang declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by V. Loia.
Rights and permissions
About this article
Cite this article
Liu, H., Geng, F., Guo, Q. et al. A fast weak-supervised pulmonary nodule segmentation method based on modified self-adaptive FCM algorithm. Soft Comput 22, 3983–3995 (2018). https://doi.org/10.1007/s00500-017-2608-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-017-2608-5