Abstract
Protein–protein interactions (PPIs) are the basis to interpret biological mechanisms of life activity, and play vital roles in the execution of various cellular processes. The development of computer technology provides a new way for the effective prediction of PPIs and greatly arouses people’s interest. The challenge of this task is that PPIs data is typically represented in high-dimensional and is likely to contain noise, which will greatly affect the performance of the classifier. In this paper, we propose a novel feature weighted rotation forest algorithm (FWRF) to solve this problem. We calculate the weight of the feature by the \(\chi ^{2}\) statistical method and remove the low weight value features according to the selection rate. With this FWRF algorithm, the proposed method can eliminate the interference of useless information and make full use of the useful features to predict the interactions among proteins. In cross-validation experiment, our method obtained excellent prediction performance with the average accuracy, precision, sensitivity, MCC and AUC of 91.91, 92.51, 91.22, 83.84 and 91.60% on the H. pylori data set. We compared our method with other existing methods and the well-known classifiers, such as SVM and original rotation forest on the H. pylori data set. In addition, in order to demonstrate the ability of the FWRF algorithm, we also verified on the Yeast data set. The experimental results show that our method is more effective and robust in predicting PPIs. As a web server, the source code, H. pylori data sets and Yeast data sets used in this article are freely available at http://202.119.201.126:8888/FWRF/.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Proteins are the most crucial macromolecules of life and participate in almost all process within the cell, including signaling cascades, DNA transcription and replication, and metabolic cycles (Yin et al. 2014). It has been confirmed that proteins rarely perform their functions independently. Instead, they cooperate with other macromolecules, and especially other proteins perform their functions by forming a huge network of protein–protein interactions (PPIs) (Gavin et al. 2002). Therefore, the study of protein–protein interactions has been the central issue in system biology (Theofilatos et al. 2011; Tuncbag et al. 2009; Zhu et al. 2011; You 2010).
In recent years, a variety of experimental techniques have been developed for the detection of protein interactions, such as Yeast two-hybrid screens (Ito et al. 2001; Ho et al. 2002; Krogan et al. 2006), protein chip (Zhu et al. 2001), mass spectrometric protein complex identification (Ho et al. 2002). These experimental methods revealed many unknown interactions. However, due to the limitations of high false positive rate, time-consuming, high cost and low coverage, the results obtained by the experimental method are far from the expectations of the researchers. Therefore, there is an urgent need to develop reliable computational methods for predicting protein–protein interactions as a complement of the experimental methods to solve these problems in the post-genomic era (Jin 2000; Guo et al. 2008; You et al. 2010, 2016; Shen et al. 2007; Zhu 2015).
At the same time, a number of computational methods have been proposed for the prediction of protein–protein interactions (Ji et al. 2014; Zhu et al. 2013a, b; Lin et al. 2013; Jin et al. 2002). These methods can be divided into the phylogenetic profile method (Pazos and Valencia 2001), the information of gene neighboring (Ideker et al. 2002), the interacting proteins coevolution method (Pazos et al. 1997), phylogenetic relationship (Pazos et al. 1997), the literature mining method (Marcotte et al. 2001), gene fusion events (Enright et al. 1999), gene co-expression (Ideker et al. 2002) and so on (Zhu et al. 2015). However, the application of these methods is limited (Yin et al. 2008; Mao et al. 2007) because these methods depend on the prior information of the protein pairs. Therefore, the method of obtaining information directly from the protein amino acid sequence has attracted more and more researchers’ interest (Guo et al. 2008; Shen et al. 2007; Zhu et al. 2015; Bock and Gough 2003; Martin et al. 2005; Zhang et al. 2012; Nanni 2005; Nanni and Lumini 2006; Gao et al. 2016; Jin and Sendhoff 2008). At present, many researchers are engaged in the development of sequence-based methods to predict protein interactions. For example, You et al. (2013) developed the principal component analysis–ensemble extreme learning machine (PCA–EELM) method uses only protein sequence information to predict PPIs. This method yields 87.00% prediction accuracy, 86.15% sensitivity and 87.59% precision when performed on the PPIs data of Saccharomyces cerevisiae. Shen et al. (2007) considered the residues local environments and developed a SVM model by combining a conjoint triad feature with S-kernel function of protein pairs to predict PPIs network. This model has yielded a high prediction accuracy of 83.93% when performed on human PPIs data set. Martin et al. (2005) proposed a model which is a product of subsequences and an expansion of the signature descriptor from chemical information to detect PPIs. The accuracy obtained by this model was 80% when using tenfold cross-validation on the Yeast data sets.
In this paper, we propose a sequence-based method to predict protein–protein interactions, which based on the feature weighted rotation forest algorithm (FWRF) (Rodriguez and Kuncheva 2006; Nanni and Lumini 2009). In particular, we convert the protein amino acid sequence into the Position-Specific Scoring Matrix (PSSM) (Gribskov et al. 1987), and then put the features which extracted by local phase quantization (LPQ) (Ojansivu and Heikkila 2008) into the feature weighted rotation forest classifier to predict PPIs. In the data set, the importance of features is inconsistent, some features contain more information, some contain less information, and even some contain interference information. If the features containing the interference information can be eliminated, the classifier will be easier to classify the samples. Based on this idea, we added the concept of weight to the feature and improved the original rotation forest algorithm. After the feature is assigned a weight, the new algorithm can remove the features of the low value, namely noise, so the classifier can obtain more accurate information from the feature subset. In the experiment, we tested the improved algorithm on different data sets, and compared with other excellent methods. The experimental results show that our feature weighted rotation forest algorithm does improve the accuracy of the classification and the efficiency of calculation.
2 Materials and methodology
2.1 Data sources
We implement our method on H. pylori data set, which introduced by Rain et al. (2001). The H. pylori data set consists of 2916 protein pairs, of which 1458 pairs are interacting and 1458 pairs are non-interacting. This data set provides a platform for comparing our method with other methods, and can be downloaded at http://www.cs.sandia.gov/~smartin/software.html.The Yeast data were extracted from S. cerevisiae core subset of database of interacting proteins (DIP) (Xenarios et al. 2002), version DIP_20070219. After removing the protein pairs with less than 50 residues and greater than 40% sequence identity, the final Yeast data set contains 5594 positive samples and 5594 negative samples.
2.2 Position-Specific Scoring Matrix (PSSM)
Position-Specific Scoring Matrix (PSSM) was produced from a set of sequences previously aligned by sequence or structural similarity. It was used to detect distantly related protein introduced by Gribskov et al. (1987). A PSSM is a matrix of \(L\times 20\), where row represents the total number of amino acids in a protein and column represents the 20 naive amino acids. Let \(\hbox {PSSM}=\left\{ {P_{i,j} :i=1\ldots L{\text { and }}j=1\ldots 20} \right\} \) and each matrix is represented as follows:
where \(P_{i,j}\) in the i row of PSSM mean that the probability of the ith residue being mutated into type j of 20 native amino acids during the procession of evolutionary in the protein from multiple sequence alignments.
In our study, we used the Position-Specific Iterated BLAST (PSI-BLAST) tool (Altschul et al. 1997) and SwissProt database on a local machine to create PSSM for each protein sequence. PSI-BLAST is a highly sensitive protein sequence alignment program, particularly effective in the discovery of new members of a protein family and similar protein of distantly related species. In order to obtain broad and high homologous sequences, we set the value of e-value is 0.001, the number of iterations is 3 and the value of other parameters are default. Applications of PSI-BLAST and SwissProt database can be downloaded at http://blast.ncbi.nlm.nih.gov/Blast.cgi.
2.3 Local phase quantization (LPQ)
Local phase quantization (LPQ) was originally described in the article (Ojansivu and Heikkila 2008) for texture description by Ojansivu and Heikkila. The LPQ method is based on the blur invariance property of the Fourier phase spectrum (Wang et al. 2015; Li et al. 2011; Li and Olson 2010). The discrete model for spatially invariant blurring of an original image f(x) apparent in an observed image g(x) can be expressed as a convolution, the formula is as follows:
where x is a vector of coordinates \(\left[ {x,y} \right] ^\mathrm{T}\), \(\times \) indicates two-dimensional convolutions and h(x) represents the point spread function (PSF) of the blur. In the Fourier domain, this is equivalent to
where u represents a vector of coordinates \(\left[ {u,v} \right] ^\mathrm{T}\), G(u), F(u) and H(u) are the discrete Fourier transforms (DFT) of the blurred image, the original images and the point spread function (PSF), respectively. Consider only the phase of the spectrum, the phase relations can be expressed as
If the spread point function h(x) is centrally symmetric, \(H\in \left\{ {0,\pi } \right\} \) as the Fourier transform H(u) is always real. So its phase can only be represented as a two-valued function:
This means that
In local phase quantization, a short-term Fourier transform (STFT) computed over a rectangular M-by-M neighborhoods \(N_x \) at each pixel position of an image f(x) is defined by:
where \(f_x \) is another vector containing all \(M^{2}\) image samples from \(N_x \), \(\omega _u \) is the basis vector of the two-dimensional DFT at frequency u.
The local Fourier coefficients are computed at four frequency points: \(u_1 =\left[ {a,0} \right] ^\mathrm{T}\), \(u_2 =\left[ {0,a} \right] ^\mathrm{T}\), \(u_3 =\left[ {a,a} \right] ^\mathrm{T}\), and \(u_4 =\left[ {a,-a} \right] ^\mathrm{T}\), where a is a sufficiently small scalar to satisfy \(H\left( {u_i} \right) >0\). So each pixel point can be expressed as a vector, given by:
Using a simple scalar quantizer, the resulting vectors are quantized:
where \(g_j (x)\) is the jth component of \(F_x \). After quantization, \(F_x \) becomes an eight-bit binary number vector, and each component is assigned a weight of \(2^{j}\). The resulting eight binary coefficients are represented as integer values between 0 and 255 using binary coding
From all these values, we obtain a histogram that can be represented as a 256-dimensional feature vector. In this article, each PSSM matrix from the H. pylori data set is converted to 256-dimensional feature vectors by using the LPQ method.
3 Feature weighted rotation forest algorithm
3.1 Original rotation forest classifier
Rotation forest (RF) is a popular ensemble classifier. In order to generate the training samples of the base classifier, the feature set is randomly divided into K subsets. The linear transformation method is applied in each subset, and retains all the principal components to maintain the precision of data. The rotation formed the training sample of new features to ensure the diversity of data. Hence the rotation forest can enhance the accuracy for individual classifier and the diversity in the ensemble at the same time. The framework of RF is described as follows.
Let X is the training sample set, Y is the corresponding labels and F is the feature set. Assuming \(\left\{ {x_i,y_i} \right\} \) contains D training samples, where in \(x_i =\left( {x_{i1},x_{i2} ,\ldots ,x_{in}} \right) \) be an n-dimensional feature vector. Then X is \(D\times n\) matrix, which is composed of n observation feature vector composition. The feature set is randomly divided into K equal subsets by a suitable factor. Let the number of decision trees is L, then the decision trees in the forest can be represented as \(T_1,T_2,\ldots ,T_L\). The implementation process of the algorithm is as follows.
-
(1)
Select the suitable parameter K which is a factor of n, let F randomly divided into K parts of the disjoint subsets, each subset contains a number of features is \(N = n/k\).
-
(2)
From the training dataset X select the corresponding column of the feature in the subset \(T_{i,j} \), form a new matrix \(X_{i,j} \). Followed by a bootstrap subset of objects extracted 3 / 4of X constituting a new training set \({X}^{\prime }_{i,j}\).
-
(3)
Matrix \({X}^{\prime }_{i,j}\) is used as the feature transform for producing the coefficients in a matrix \(P_{i,j} \), which jth column coefficient as the characteristic component jth.
-
(4)
The coefficients obtained in the matrix \(P_{i,j} \) are constructed a sparse rotation matrix \(S_i \), which is expressed as follows:
$$\begin{aligned} S_i =\left[ \begin{array}{c@{\quad }c@{\quad }c@{\quad }c} f_{i,1}^{(1)},\ldots , f_{i,1}^{({N_1})} &{}0 &{} \cdots &{}0 \\ 0 &{}f_{i,2}^{(1)},\cdots ,f_{i,2}^{({N_2})} &{} \cdots &{}0 \\ \vdots &{}\vdots &{} \ddots &{}\vdots \\ 0&{}0&{}\cdots &{} f_{i,k}^{(1)},\ldots ,f_{i,k}^{({N_k})} \\ \end{array}\right] \nonumber \\ \end{aligned}$$(11)
In the prediction period, provided the test sample x, generated by the classifier \(T_i \) of \(d_{i,j} \left( {XS_i^f} \right) \) to determine x belongs to class \(y_i \). Next, the class of confidence is calculated by means of the average combination, and the formula is as follows:
Then assign the category with the largest \(\alpha _j (x)\) value to x.
3.2 Improved rotation forest with weighted feature selection
With the rapid development of technology, we get more and more features, the feature dimension is also more and more high. The high-dimensional data contain more information and reflect the fact more comprehensive. However, with the increase in dimensions, redundant information is also increasing. In view of how to effectively analyze and integrate information, extract useful information from a large amount of noise data, combined with feature selection technology, we improved the rotation forest algorithm. We use \({\chi }^{2}\) statistical method to calculate the weight of the features. A feature F against the class feature is computed using the following formula:
where n is the number of values in feature F, \(u_{ij} \) is the count of the value \(\lambda _i \) in feature F and belong to class \(c_j \), defined as:
\(v_{i,j} \) is the expected value of \(\lambda _i \) and \(c_j \), defined as:
where \(\hbox {count}\left( {F=\lambda _i} \right) \) is the number of samples in the feature F value is \(\lambda _i \), \(\hbox {count}\left( {C=c_j} \right) \) is the number of samples in the class C value is \(c_j\), and N is the total number of samples in the training set.
In order to make full use of the useful information, we perform the following steps. Firstly, calculate the weight of each feature by the formula 13; secondly, sorts the features in descending order according to the weight value; finally, select new features from the full feature set in accordance with a given feature selection rate r. After these steps, we construct a new data set for the algorithm. Using the new data set can not only effectively eliminate the noise, especially the high-dimensional data, but also can reduce the computation time.
4 Results and discussion
4.1 Selection of the number of features
In the experiment, fivefold cross-validation is used to evaluate the performance of our method. The whole data set is randomly split into 5 equal-sized subsets. In order to ensure the independence of the data in the experiment, we select one subset as the test set, and the other four subsets as the training set. Loop 5 times in this way, such that each subset is used for testing exactly once. In our method, the number of features is controlled by the feature selection rate r. Different feature selection rates will lead to different accuracy and computation time. We take one subset as the test set, and the rest for the training set to experiment when selecting the parameters. The range of value r is from 0 to 1. In order to prevent the loss of too much information, in the experiment we test the value of \(r = 0.2, 0.3, \ldots , 1.0\). Figure 1 plots the influence of feature selection on classifier performance and Fig. 2 plots the influence of feature selection on time cost.
Figure 1 shows that the accuracy increases slowly as r increases when the feature selection rate r is less than 0.3. The reason is that the number of features increases with increasing r, and more information is provided to the classifier. The accuracy reaches its peak when the rate is 0.4. This means that the feature set has contained the most useful information. The accuracy gradually decreases with increasing rates when the rate is greater than 0.4. This proves that the high-dimensional data are likely to contain noise. It can be seen from the figure that \(r=0.4\) is the best setting of this experiment.
Figure 2 shows the influence of feature selection on time cost. It is obvious from the figure that with the increase in the feature select rate, namely the increase in the number of features, the computation time continues to rise. This indicates that the dimension of the data on the impact of computing time is very large.
4.2 Select the parameters of the FWRF classifier
Another problem affecting the performance of our model is the number of decision trees L and the number of feature subset K in the feature weighted rotation forest. Since a large number of trees and feature subsets will lead to considerable computational cost and will affect the accuracy of the improved, we need to find the most suitable parameters by the grid search method. Figure 3 shows the predicted results of different parameters. We can see that with the increase of L, the accuracy rate increases rapidly at the beginning, and then tends to be gentle. However, there is no significant change in accuracy as K increased. Considering the time cost and accuracy of the algorithm, we finally select the most appropriate parameters \(K=3\) and \(L=53\).
4.3 Assessment of prediction ability
In the experiment, the evaluation criteria are reflected by the prediction accuracy (Accu.), sensitivity (Sen.), precision (Prec.), and Matthews correlation coefficient (MCC). The calculation formulas are listed below:
where TP, TN, FP, FN represent the number of true positives, true negatives, false positives and false negatives, respectively. Besides, the receiver operating characteristic (ROC) curve (Zweig and Campbell 1993) and the area of the ROC curve (AUC) are employed to visually show the performance of classifier.
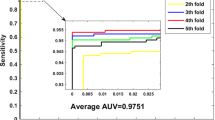
Our prediction results are shown in Table 1. We can see that the average accuracy, precision, sensitivity, MCC and AUC of 91.91, 92.51, 91.22, 83.84 and 91.60%, respectively. The standard deviations of them are 1.21, 1.83, 1.92, 2.44 and 0.55%, respectively. The ROC curves performed on H. pylori data set was shown in Fig. 4. In this figure, X-ray depicts false positive rate (FPR) while Y-ray depicts true positive rate (TPR).
4.4 Comparison with original RF and SVM
To evaluate the performance of our method, we compared with the original rotation forest classifier and the state-of-the-art SVM classifier. In the comparison, we use the same feature extraction method and implement on the same data set. After optimization of the grid search method, the parameters K and L of the original rotation forest are set to 2 and 3, the parameters c and g of the SVM are set to 0.08 and 22. Table 2 lists the experimental results of the original rotation forest classifier. From the table we can see that the average accuracy, precision, sensitivity, MCC, AUC and their standard deviations of \(85.36 \pm 1.61\), \(85.29 \pm 2.00\), \(85.37 \pm 3.01\), \(70.72 \pm 3.26\) and \(85.61 \pm 2.67\)%, respectively. Table 3 lists the experimental results of the SVM classifier. From the table we can see that the average accuracy, precision, sensitivity, MCC, AUC and their standard deviations of \(81.82 \pm 2.19\), \(82.82 \pm 4.15\), \(80.29 \pm 1.03\), \(63.71 \pm 4.48\) and \(88.83 \pm 1.94\)%, respectively.
From the table it can be seen that the performance of our FWRF showed significant improvement over the other two classifiers. The average accuracy is 6.55% higher than original RF, and 10.09% higher than SVM. This was due to the fact that there may contain a lot of noise information in the feature set, which will affect the accuracy of the classifier, so the original RF and SVM did not perform well on this kind of feature set. In addition, the processed feature set is likely to be smaller than the original feature set, which reduces the dimension of the data, so it can reduce the running time of the classifier and improve efficiency (Figs. 5, 6).
4.5 Comparison of the proposed method with other existing methods
Many methods have been proposed for predicting protein interactions, and good results have been obtained. In this section, we focused on the H. pylori data set to compare the FWRF with other algorithms. Table 4 lists the average prediction results of the other six different methods on the H. pylori data set. We can see that the accuracy values obtained by these methods are between 75.8 and 87.50%. Their average accuracy rate is 82.80, 9.11% lower than ours. The average precision, sensitivity and MCC values of these methods are lower than our method, which are 83.79, 81.95, 74.38%, respectively.
4.6 Performance of the feature weighted rotation forest on Yeast data set
In order to further evaluate the performance of the feature weighted rotation forest classifier, we verified it on Yeast data set. In the experiment, the features of the Yeast data set are also extracted by the LPQ algorithm. Figure 7 shows the experimental results for the different selection ratios of the feature weighted rotation forest classifier. From the figure we can see that the feature weighted rotation forest classifier performs well. The accuracy, precision specificity and MCC would be increased along with the increasing of the selection ratio when the selection ratio less than 90%; the peak is reached when the selection ratio is 90%; and decreased when the selection ratio more than 90%. This experiment demonstrates that our improved algorithm is equally applicable to Yeast data set. In addition, we can see that the features extracted by the LPQ algorithm in the Yeast data set contain less noise than in the H. pylori data set.
5 Conclusion
In this paper, we propose a novel improved rotation forest algorithm based on weighted feature selection strategy to predict the interactions among proteins. The improved algorithm considers that the high-dimensional data of PPIs may contain noise. To solve this problem, we distinguish the importance of different features by weight, and delete the features with small weight according to the selection ratio, that is, the noise features. This strategy can improve the accuracy of the classifier while reducing the dimension of the data and saving the execution time. In the experiment, we verify its capability on H. pylori and Yeast data set, and compare it with original rotation forest classifier, SVM classifier and other existing methods. Excellent experimental results show that our method is effective and efficient. In future research, we intend to apply the feature weighted rotation forest algorithm in more areas and look forward to gooderformance.
References
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–402
Bock JR, Gough DA (2003) Whole-proteome interaction mining. Bioinformatics 19(1):125–134
Enright AJ, Iliopoulos I, Kyrpides NC et al (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature 402(6757):86–90
Gao ZG, Wang L, Xia SX et al (2016) Ens-PPI: a novel ensemble classifier for predicting the interactions of proteins using autocovariance transformation from PSSM. Biomed Res Int 2016(4):1–8
Gavin AC, Bosche M, Krause R et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868):141–147
Gribskov M, McLachlan AD, Eisenberg D (1987) Profile analysis: detection of distantly related proteins. Proc Nat Acad Sci USA 84(13):4355–8
Guo Y, Yu L, Wen Z et al (2008) Using support vector machine combined with auto covariance to predict protein–protein interactions from protein sequences. Nucleic Acids Res 36(9):3025–3030
Ho Y, Gruhler A, Heilbut A et al (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415(6868):180–183
Ideker T, Ozier O, Schwikowski B et al (2002) Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics (Oxford, England) 18 Suppl 1:S233-40
Ito T, Chiba T, Ozawa R et al (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Nat Acad Sci USA 98(8):4569–4574
Ji Z, Wang B, Deng S et al (2014) Predicting dynamic deformation of retaining structure by LSSVR-based time series method. Neurocomputing 137:165–172
Jin Y (2000) Fuzzy modeling of high-dimensional systems: complexity reduction and interpretability improvement. IEEE Trans Fuzzy Syst 8(2):212–221
Jin Y, Sendhoff B (2008) Pareto-based multiobjective machine learning: an overview and case studies. IEEE Trans Syst Man Cybern Part C 38(3):397–415
Jin Y, Olhofer M, Sendhoff B (2002) A framework for evolutionary optimization with approximate fitness functions. IEEE Trans Evol Comput 6(5):481–494
Krogan NJ, Cagney G, Yu HY et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440(7084):637–643
Li Y, Olson EB (2010) A general purpose feature extractor for light detection and ranging data. Sensors 10(11):10356–10375
Li Y, Olson EB, IEEE (2011) Structure tensors for general purpose LIDAR feature extraction. In: IEEE international conference on robotics and automation ICRA, pp 1869–1874
Lin Z, You ZH, Huang DS et al (2013) t-LSE: a novel robust geometric approach for modeling protein-protein interaction networks. Plos One 8(4):e58368
Liu B, Yi J, Aishwarya SV et al (2013) QChIPat: a quantitative method to identify distinct binding patterns for two biological ChIP-seq samples in different experimental conditions. BMC Genom 14(8):S3
Mao Y, Xia Z, Yin Z et al (2007) Fault diagnosis based on fuzzy support vector machine with parameter tuning and feature selection. Chin J Chem Eng 15(2):233–239
Marcotte EM, Xenarios I, Eisenberg D (2001) Mining literature for protein–protein interactions. Bioinformatics 17(4):359–363
Martin S, Roe D, Faulon JL (2005) Predicting protein–protein interactions using signature products. Bioinformatics 21(2):218–226
Nanni L (2005) Hyperplanes for predicting protein–protein interactions. Neurocomputing 69(1–3):257–263
Nanni L, Lumini A (2006) An ensemble of K-local hyperplanes for predicting protein–protein interactions. Bioinformatics 22(10):1207–1210
Nanni L, Lumini A (2009) Ensemble generation and feature selection for the identification of students with learning disabilities. Expert Syst Appl 36(2):3896–3900
Ojansivu V, Heikkila J (2008) Blur insensitive texture classification using local phase quantization. Image Signal Process 5099:236–243
Pazos F, Valencia A (2001) Similarity of phylogenetic trees as indicator of protein–protein interaction. Protein Eng 14(9):609–614
Pazos F, Helmer-Citterich M, Ausiello G et al (1997) Correlated mutations contain information about protein–protein interaction. J Mol Biol 271(4):511–523
Rain JC, Selig L, De Reuse H et al (2001) The protein–protein interaction map of Helicobacter pylori (vol 409, pg 211, 2001). Nature 409(6821):743
Rodriguez JJ, Kuncheva LI (2006) Rotation forest: a new classifier ensemble method. IEEE Trans Pattern Anal Mach Intell 28(10):1619–1630
Shen J, Zhang J, Luo X et al (2007) Predictina protein–protein interactions based only on sequences information. Proc Nat Acad Sci USA 104(11):4337–4341
Theofilatos KA, Dimitrakopoulos CM, Tsakalidis AK et al (2011) Computational approaches for the prediction of protein–protein interactions: a survey. Curr Bioinform 6(4):398–414
Tuncbag N, Kar G, Keskin O et al (2009) A survey of available tools and web servers for analysis of protein–protein interactions and interfaces. Brief Bioinform 10(3):217–232
Wang H, Song A, Li B et al (2015) Psychophysiological classification and experiment study for spontaneous EEG based on two novel mental tasks. Technol Health Care 23:S249–S262
Xenarios I, Salwinski L, Duan XQJ et al (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res 30(1):303–305
Yin Z, Zhou X, Bakal C et al (2008) Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinform 9(1):264
Yin Z, Deng T, Peterson LE et al (2014) Transcriptome analysis of human adipocytes implicates the NOD-like receptor pathway in obesity-induced adipose inflammation. Mol Cell Endocrinol 394(1–2):80–87
You ZH (2010) Using manifold embedding for assessing and predicting protein interactions from high-throughput experimental data. Bioinformatics 26(21):2744–2751
You ZH, Yin Z, Han K et al (2010) A semi-supervised learning approach to predict synthetic genetic interactions by combining functional and topological properties of functional gene network. BMC Bioinform 11(1):343
You ZH, Lei YK, Zhu L et al (2013) Prediction of protein-protein interactions from amino acid sequences with ensemble extreme learning machines and principal component analysis. BMC Bioinform 14(8):1–11
You ZH, Zhou M, Luo X et al (2016) Highly efficient framework for predicting interactions between proteins. IEEE Trans Cyber 1–13
Zhang YQ, Zhang DL, Mi G et al (2012) Using ensemble methods to deal with imbalanced data in predicting protein–protein interactions. Comput Biol Chem 36:36–41
Zhu Z (2015) CompMap: a reference-based compression program to speed up read mapping to related reference sequences. Bioinformatics 31(3):426–8
Zhu H, Bilgin M, Bangham R et al (2001) Global analysis of protein activities using proteome chips. Science 293(5537):2101–2105
Zhu Z, Zhou J, Ji Z et al (2011) DNA sequence compression using adaptive particle swarm optimization-based memetic algorithm. IEEE Trans Evol Comput 15(5):643–658
Zhu Z, Zhang Y, Ji Z et al (2013a) High-throughput DNA sequence data compression. Brief Bioinform 16(1):1–15. doi:10.1093/bib/bbt087
Zhu L, You Z-H, Huang D-S (2013b) Increasing the reliability of protein-protein interaction networks via non-convex semantic embedding. Neurocomputing 121:99–107
Zhu Z, Jia S, He S et al (2015) Three-dimensional Gabor feature extraction for hyperspectral imagery classification using a memetic framework. Inf Sci 298(C):274–287
Zweig MH, Campbell G (1993) Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39(4):561–77
Acknowledgements
This work is supported by the Fundamental Research Funds for the Central Universities (2017XKQY083).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
Additional information
Communicated by V. Loia.
Rights and permissions
About this article
Cite this article
Wang, L., You, ZH., Xia, SX. et al. An improved efficient rotation forest algorithm to predict the interactions among proteins. Soft Comput 22, 3373–3381 (2018). https://doi.org/10.1007/s00500-017-2582-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-017-2582-y