Abstract
MicroRNAs (miRNAs) are small RNAs that regulate genes involved in various aspects of plant development, but their presence and expression patterns in the male gametophytes of gymnosperms have not yet been established. Therefore, this study identified and compared the expression patterns of conserved miRNAs from two stages of the male gametophyte of loblolly pine (Pinus taeda), which are the mature (ungerminated) and germinated pollen. Microarray was used to identify conserved miRNAs that varied in expression between these two stages of the loblolly pine male gametophyte. Forty-seven conserved miRNAs showed significantly different expression levels between mature and germinated loblolly pine pollen. In particular, miRNAs representing 14 and 8 families were up- and down-regulated in germinated loblolly pine pollen, respectively. qRT-PCR was used to validate their expression patterns using representative miRNAs. Target genes and proteins were identified using psRNATarget program. Predicted targets of the 22 miRNA families belong mostly to classes of genes involved in defense/stress response, metabolism, regulation, and signaling. qRT-PCR was also used to validate the expression patterns of representative target genes. This study shows that conserved miRNAs are expressed in mature and germinated loblolly pine pollen. Many of these miRNAs are differentially expressed, which indicates that the two stages of the male gametophyte examined are regulated at the miRNA level. This study also expands our knowledge of the male gametophytes of seed plants by providing insights on some similarities and differences in the types and expression patterns of conserved miRNAs between loblolly pine with those of rice and Arabidopsis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) make up a minute portion of the genome, but play important roles in post-transcriptional gene regulation through cleavage of target mRNAs or translational repression (Brodersen et al. 2008; Carthew and Sontheimer 2009). Plant miRNAs have a remarkable propensity to target genes involved in developmental processes, phase transitions, metabolism, and defense and stress responses (Jones-Rhoades et al. 2006; Mallory and Vaucheret 2006; Chen 2009; Lelandais-Briere et al. 2010). Many miRNAs are deeply conserved in land plants (Floyd and Bowman 2004; Axtell and Bartel 2005; Kidner and Martienssen 2005), but many have been found to be taxa, tissue, or stage specific (Arazi et al. 2005; Lu et al. 2005; Zhang et al. 2006a, b, 2010; Zhan and Lukens 2010). All these support the notion that the regulation of conserved miRNA activity at various stages of growth and in specific cell types is of central importance for normal plant development. In seed plants, our knowledge of miRNAs is mostly from reports on structures representing the sporophyte (diploid) phase of the life cycle (e.g., embryos, seedlings, leaves, stems, wood, etc.) (Floyd and Bowman 2004; Axtell and Bartel 2005; Jones-Rhoades et al. 2006; Lu et al. 2007; Chen 2009; Zhao et al. 2010; Zhang et al. 2012). Several studies have also demonstrated miRNA expression from the male gametophyte (haploid) phase in various species of angiosperms (Fujioka et al. 2008; Chamber and Shuang 2009; Grant-Downton et al. 2009; Grant-Downton 2010; Le Trionnaire and Twell 2010; Borges et al. 2011; Le Trionnaire et al. 2011; Wei et al. 2011; Peng et al. 2012), but none from any gymnosperm.
In seed plants, the male gametophyte phase produces the mature pollen grain which is typically formed in the microsporangia in the pollen cones of gymnosperms and anthers in the flowers of angiosperms. It becomes dispersed and typically ends up in the ovule (in gymnosperms) or stigma (in angiosperms) where it germinates and forms a pollen tube that generally functions to deliver the sperm to the egg. In conifers, mature pollen grains are dispersed at one- to five-cell stage, except in Podocarpaceae where the prothallial cells may proliferate and so their pollen grains may be shed with as many as 40 nuclei (Fernando et al. 2010). Pine pollen grains are typically composed of five cells at the time that they are released from the pollen cones. Three of these cells (two prothallial cells and a sterile cell) have no known function in post-pollination processes, whereas the vegetative and generative cells are involved in pollen tube growth and gamete formation, respectively. Pine pollen germinates on the nucellus of the ovule within 5 days from pollination, and the pollen tube grows to about half of the thickness of the nucellus and then ceases to elongate. The pollen tube remains dormant for about a year inside the ovule with the generative cell remaining undivided. Therefore, much of the post-pollination processes in pines and conifers in general are involved in pollen tube growth. There are many other major morphological and developmental differences between the male gametophytes of gymnosperms and angiosperms (Singh 1978; Gifford and Foster 1989; Fernando et al. 2010; Williams 2012), but it remains to be seen whether these are accompanied by major differences at the molecular level. Nevertheless, proper regulation of gene expression is necessary in the transition of the male gametophyte from the mature (ungerminated) to germinated phase, two stages that are spatially and temporally separated in vivo and represent pre-pollination and post-pollination stages, respectively. As such, many transcripts and proteins have been shown to be differentially expressed between mature and germinated pollen in gymnosperms (Fernando 2005; Chen et al. 2006) and angiosperms (Dai et al. 2007; Zou et al. 2009; Zhu et al. 2011) and likely include the miRNAs that are involved in modulating these two stages. In angiosperms, many miRNAs show dynamic and complex changes related to the development of microspores to mature pollen with expressions that are distinct from those of the sporophytic tissues (Fujioka et al. 2008; Chambers and Shuai 2009; Grant-Downton et al. 2009; Borges et al. 2011; Le Trionnaire et al. 2011; Wei et al. 2011; Peng et al. 2012). MiRNAs that are associated with bicellular and tricellular pollen of rice have also been described, and many differences between their miRNA profiles have been found (Peng et al. 2012). Thus far, our knowledge of spatiotemporal difference in miRNA expression during male gametophyte development in seed plants is confined to pre-pollination stages. Therefore, much remains to be discovered regarding the miRNAs in the male gametophyte phase, particularly those that are related to the post-pollination stages such as the transition from mature to germinated phase.
There is limited information about miRNA analysis in gymnosperms and almost entirely based on sporophytic tissues from conifers. Lu et al. (2007) identified miRNAs from loblolly pine (Pinus taeda) stem xylem and examined their expressions in relation to fusiform gall development. Qiu et al. (2009) examined the miRNAs that are up-regulated in a Taxus chinesis somatic cell line upon exposure to methyl jasmonate. Using Norway spruce (Picea abies) seedlings, novel and conserved miRNAs were identified, but most of which targeted unknown genes (Yakovlev et al. 2010). In Japanese larch (Larix leptolepis), four miRNA families are differentially expressed between embryonic and non-embryonic callus (Zhang et al. 2010). Stage-specific expression of nine representative miRNAs and their targets demonstrated their potential in modulating somatic embryogenesis in Japanese larch (Zhang et al. 2012). A remarkable finding regarding large-scale comparison of miRNAs in gymnosperms (including Cycas, Ginkgo, and Ephedra but mostly conifers) versus angiosperms is the prevalence of 21nt long small RNAs, which has been considered as a gymnosperm-unique silencing signature and is believed to be related to specific Dicer-like family genes in gymnosperms (Dolgosheina et al. 2008; Morin et al. 2008; Yakovlev et al. 2010). However, recent reports indicate that this size bias is associated only with vegetative organs and tissues since reproductive structures show appreciable amounts of 24nt long small RNAs (Wan et al. 2012a; Nystedt et al. 2013; Zhang et al. 2013). The only report on gametophytic tissue expression in gymnosperm is that of Oh et al. (2008), which compared the expression of five conserved miRNAs between the female gametophyte and zygotic embryo of loblolly pine. To fill the gap in our knowledge of miRNA expression in the male gametophytes in gymnosperms, this study aimed to identify conserved miRNAs from mature and germinated pollen of loblolly pine through microarray analysis, validate their expression patterns through qRT-PCR, and predict their target genes and functions using web-based computational approaches.
Materials and methods
Collection and in vitro germination of pollen grains
Pollen cones were collected from several loblolly pines (P. taeda) growing in the campus of North Carolina State University, Raleigh, NC, 1–2 days before pollen grains were released from the microsporangia. The pollen grains at this stage are five-celled and are in the process of dehydration in preparation for dormancy and dispersal. The pollen cones per individual tree were labeled and separately processed for surface sterilization following Fernando et al. (1997). Prior to storage of the sterile pollen grains at −80 °C, average pollen germination was determined to be 98 %. Total RNA was isolated from the frozen pollen grains to represent the mature pollen used in the study. For germinated pollen, stored pollen grains were evenly dispensed on modified Brewbaker and Kwack (1963) medium (pH 5.8) composed of 0.1 mg ml−1 H3BO3, 0.3 mg ml−1 Ca2NO3, 0.2 mg ml−1 MgSO4, and 0.1 mg ml−1 KNO3, supplemented with 145 mM sucrose and 0.4 % phytagel (Fernando et al. 1997). A 25-mm nitrocellulose membrane (Millipore, Billerica, MA) was placed on the surface of the solidified germination medium, and 30 mg pollen grains were dispensed onto the membrane by gently shaking a sterile spatula containing pollen grains over it. The culture plates were incubated at 25 °C in the dark for 2 days (48 h) to allow pollen grains to germinate. Pollen grains were considered germinated if the pollen tube was at least as long as the diameter of the pollen grain. Average pollen germination was 98 %. DAPI staining indicates that both mature and germinated pollen grains are composed of five nuclei.
RNA preparation, miRNA array, and data analysis
Using pollen grains from three different loblolly pine individuals as biological replicates, total RNA was extracted from mature (directly from frozen and stored pollen grains) and germinated (2-day-old pollen cultures) pollen using TRIzol Reagent (Invitrogen, Carlsbad, CA). The three biological samples from mature pollen were designated as U1, U2, and U3 and for germinated pollen as G1, G2, and G3. Total RNAs from these six samples were sent to LC Sciences Inc. (Houston, TX) for the miRNA array procedure. Briefly, this involved the use of a custom μparaflo™ microfluidic array containing MiRBase (Release 10.1) plant miRNA probes. The quality of the total RNA was verified, and the samples were size fractionated to isolate the small RNAs (<300nt), to which 3′ poly (A) tails were added. An oligonucleotide tag was ligated to the poly (A) tail for use in fluorescent dye staining. The 20mer RNA controls, PUC2PM-20B and PUC2MM-20B, were spiked into the small RNA samples for use in quality control during the miRNA array assay. The small RNAs were then labeled with Cy3 or Cy5 fluorescent dyes and hybridized to a dual-channel μparaflo™ microfluidics chip. The chip contained probes of chemically modified nucleotide-coding segments complementary to the 653 unique plant miRNA sequences from MiRBase (Release 10.1), representing miRNAs from 17 plant species. For quality control, the chip also included probes for maize 5S rRNA, an inner positive control, and PUC2-20B (an artificial non-homologous nucleic acid), an external positive control. Blank and non-homologous nucleic acids were also used as negative controls. Following hybridization, images from the chip were collected with a laser scanner and digitized using Array-Pro image analysis software (Media Cybernetics, Silver Springs, MD). The signal values from the miRNA array were corrected by subtracting the background using a local regression method. To remove any system-related signal variations, the data were also normalized to the statistical mean of all detectable transcripts using a cyclic locally weighted regression (LOWESS) filter following background subtraction. The log-transformed ratios of the detected signals for mature and germinated samples were calculated along with the p values of the t test. Only miRNAs with log ratios of one or more and negative one or less (which were equal to fold changes of two or greater) and p values of equal to or less than 0.01 were considered as differentially expressed.
qRT-PCR and data analysis
The expressions of 12 representative differentially expressed conserved miRNAs representing 12 different families were validated through qRT-PCR using the NCode SYBR GreenER miRNA qRT-PCR kit (Invitrogen, Carlsbad, CA). The specific miRNAs used to represent the family were chosen generally, based on those that showed the highest and lowest expression levels from the microarray analysis. For each miRNA and the internal reference (Supplement 1), three biological replicates were performed for both mature (U1, U2, and U3) and germinated (G1, G2, and G3) loblolly pine pollen. Sequence-specific primers were used (Supplement 1). Primer specific for P. taeda 5S rRNA was used as the reference for internal normalization for comparative C T analyses. All qRT-PCRs were performed in a MiniOpticon Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Results were viewed using the CFX Manager Software (Bio-Rad, Hercules, CA) and exported to Microsoft Excel to calculate the mean C T, ΔC T, ΔΔC T, and fold change. The fold change was calculated as \(2^{ - \varDelta \varDelta C}\tau\), where ΔΔC T = ΔC T germinated pollen −ΔC T mature pollen and ΔC T = C T target miRNA −C T of reference RNA. A fold change ≥1 indicated an increased expression of the miRNA in germinated pollen as compared to mature pollen. For fold change values <1, the negative inverse was calculated to indicate the fold change reduction in expression in germinated pollen. A fold change of −1.0 or lower indicated decreased miRNA expression in germinated pollen as compared to mature pollen.
Target prediction for conserved miRNAs
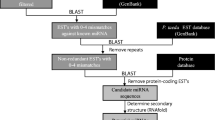
Mature miRNAs sequences from each of the representative miRNAs were retrieved from miRBase (http://www.sanger.ac.uk/Software/Rfam/mirna/). Their potential targets were determined by searching for complementary hits using the psRNATarget program (http://plantgrn.noble.org/psRNATarget/) with default parameters (Dai and Zhao 2011) using the Pinus (Pine) DFCI Gene Index version 9 as the source of sequence information. The psRNATarget program used the scoring scheme of miRU by Zhang (2005) where the “maximum expectation” was considered as the threshold of the score. The miRNA and target pair were discarded if the score was greater than the threshold, which was set at a default value of 3.0. More stringent cutoff threshold was 0–2.0 for lower false-positive prediction, and although not used in this study, more relaxed cutoff threshold was 4.0–5.0 for higher prediction coverage according to Zhang (2005). The psRNATarget program also provided accession numbers of target genes and identity of the target proteins based on complementary matches with other species. Identities of target proteins presented in Tables 3 and 4 were limited to land plant species when available.
Validation of potential target genes by qRT-PCR
Total RNA was isolated from mature and germinated loblolly pine pollen and from three different individuals, as described above. cDNA synthesis was done using QuantiTect Reverse Transcription Kit following the manufacturer’s protocol (Qiagen, Valencia, CA). Primers were designed for the potential targets of pta-miR166 and pta-miR951 based specifically on sequences derived from the Plant Gene Index (http://compbio.dfci.harvard.edu/tgi/tgipage.html), which were TC189536 and DR054135, respectively (Tables 3, 4). Primers for TC189536 are 5′-GATGCTAGCCCCGCTGGA-3′ and 5′-TGCTGGAGGCACATTCTGTAA-3′; primers for DR054135 are 5′-ACAATTGGCCTTTCATCTGC-3′ and 5′-TAAGTTCTGCCCCTCCTCCT-3′. The 18s rRNA was used as the reference gene (using 5′-CATGGCCGTTCTTAGTTGGT-3′ and 5′-GAGTTGATGACACGCGCTTA-3′ as left and right primers, respectively). qRT-PCR was done in triplicate for each combination of primers and cDNA templates including the reference gene using QuantiFast SYBR Green PCR Kit (Qiagen, Valencia, CA). Calculations for the relative expression levels were as described above.
Results
Differentially expressed conserved miRNAs
MiRNA array analysis revealed that of the 653 unique land plant miRNA sequences examined, 208 were detected in both mature and germinated loblolly pine pollen, but the expression levels of most of these were not significantly different. However, the expressions of 47 individual miRNAs showed significant changes between mature and germinated pollen (Table 1). Of this, 28 individual miRNAs were up-regulated in germinated pollen compared to mature pollen with fold changes that ranged from 2.00 to 6.3 (Table 1). On the other hand, 19 individual miRNAs were down-regulated in germinated pollen as compared to mature pollen with fold change reductions that ranged from −2.0 to −3.9 (Table 1). The expressions of 12 individual miRNAs representing 12 different families were verified through qRT-PCR (Table 2). The results showed the same miRNAs that were up-regulated and down-regulated as those obtained through microarray analysis, with fold change values of 1.6–14.3 and −1.5 to −3.4, respectively (Table 2). The 47 differentially expressed conserved miRNAs represent a total of 22 families, where 14 families were up-regulated (Tables 1, 3), while eight families were down-regulated (Tables 1, 4).
Potential targets of conserved miRNAs
To gain insights on the target genes and functions of the conserved miRNAs which were differentially expressed in the male gametophyte of loblolly pine, the web-based program psRNATarget was utilized to predict their potential targets using the Pinus DFCI Gene Index as source of mRNA sequences. For the 28 up-regulated individual miRNAs, which represent 14 families, 54 target genes were predicted (Table 3). Although information on the identities of target proteins and functions were not known for all the target genes, target protein identity was available for at least one of the target genes from most of the miRNA families, except for miR901, miR903, and miR1125 (Table 3). For 19 down-regulated individual miRNAs, which represent eight families, 32 target genes were predicted (Table 4). Information on the identities of target genes and functions were available for all of the families (Table 4). For both types of differentially expressed miRNAs, the majority of the functions were related to defense/stress response, followed by metabolism, gene regulation, and signaling. Two of the potential targets were verified through qRT-PCR, and the results showed that their expression patterns were inversely proportional with those of their corresponding miRNAs (Fig. 1).
Expression patterns of potential target genes of representative differentially expressed miRNAs (TC189536 and DR054135 are sequences obtained from the Pine Gene Index, which are the potential targets of pta-miR166 and pta-miR951, respectively. TC189536 encodes a Class III HD-Zip protein, whereas DR054135 encodes Toll/interleukin-1 receptor/P-LOOP/LRR protein)
Discussion
This study has revealed 47 conserved miRNAs representing 22 families that are differentially expressed between the two stages of the male gametophyte of loblolly pine that have been examined. The change in the expressions of these miRNAs highlights their dynamic nature and implies their involvement in the transition of the male gametophyte from mature (ungerminated) to germinated phase. Many of the 22 miRNA families are also expressed in the mature pollen of rice and/or Arabidopsis including miR156/157, miR162, miR166, miR168, miR169, miR171, miR390, miR395, miR396, miR397, miR844, and miR854 (Chambers and Shuai 2009; Grant-Downton 2010; Le Trionnaire and Twell 2010; Borges et al. 2011; Le Trionnaire et al. 2011; Peng et al. 2012). Interestingly, miR156/157 and miR168, which have been shown to be two of the most highly expressed miRNAs in the mature pollen of rice and Arabidopsis (Chambers and Shuai 2009; Grant-Downton et al. 2009; Borges et al. 2011; Le Trionnaire et al. 2011), are also the two most highly expressed miRNAs in the mature loblolly pine pollen. Whereas the level of the expression of miR156/157 and miR168 is not known in germinated rice or Arabidopsis pollen, these two miRNAs and miR844 showed reduced expression in germinated loblolly pine pollen. This implies that in loblolly pine, these three miRNAs exert greater roles in the formation of the mature pollen than in germinated pollen. Beside these three miRNAs, the eight other miRNAs indicated above are all up-regulated in germinated loblolly pine pollen. Interestingly, the expression pattern of the target gene of one of these, i.e., pta-miR166, which encodes a Class III HD-Zip protein, was inversely proportional to the expression pattern of its miRNA. The expression patterns of these eight miRNAs in germinated rice and/or Arabidopsis are not known. Our results show that conserved miRNAs expressed in mature pollen are also expressed in germinated pollen, although their levels of expression vary. Nevertheless, most of those identified in this study show higher expression levels in germinated pollen.
It is worth noting that all of the miRNA families indicated above are also expressed in various sporophytic tissues from several species of gymnosperms (particularly conifers) (Qiu et al. 2009; Yakovlev et al. 2010) and angiosperms (Zhang et al. 2006a, b; Chambers and Shuai 2009). MiR166 and miR171 are also expressed in loblolly pine zygotic embryos and megagametophytes (Oh et al. 2008). Collectively, these results imply that the miRNAs detected in this study are not only conserved among species but also between organs and tissues representing the sporophyte and gametophyte phases, and at various stages of development. As suggested by Wei et al. (2011), conserved miRNAs could serve as integral regulators of both sporophytic and gametophytic development. On the other hand, there are several differentially expressed conserved miRNAs reported here for loblolly pine male gametophyte (i.e., miR474, miR482, miR901, miR903, miR946, miR947, miR950, miR951, miR1026, and miR1125) that have not yet been reported in rice or Arabidopsis pollen. Whereas all of these are also expressed in sporophytic tissues (needles and stems) of various conifers (Lu et al. 2007; Morin et al. 2008; Yakovlev et al. 2010; Wan et al. 2012b), only miR947 and miR951 (up- and down-regulated in germinated loblolly pine pollen, respectively) have not yet been reported from any sporophytic tissue in angiosperm. Therefore, our results at least suggest differences and similarities in the conserved miRNA profiles of the male gametophytes from the representative gymnosperms and angiosperms. Although more detailed analysis is necessary to confirm this generalization, the large-scale miRNA analysis by Peng et al. (2012) has already pointed out several differences in the types and expression patterns in the male gametophytes of rice and Arabidopsis, both in terms of conserved and recently evolved miRNAs. Considering the many morphological and developmental differences in the male gametophytes of gymnosperms and angiosperms (Singh 1978; Gifford and Foster 1989; Fernando et al. 2010; Williams 2012), detailed comparative miRNA analysis will likely provide further differentiation between these two major groups of pollen-bearing plants. This will likely include miRNAs that relate to sperm formation since this process occurs in the formation of the mature pollen of rice and Arabidopsis, but not in the mature and germinated pine pollen until after about a year of pollen tube growth in the ovule (Fernando et al. 2010). As alluded to above, many recently evolved miRNAs will also serve to further differentiate the male gametophytes of gymnosperms and angiosperms.
The targets of the miRNAs identified in this study need to be experimentally verified, and thus, discussion on this subject has to be limited since functional analysis in gymnosperms is not feasible at the moment. With this limitation, computational prediction provides insights on the target genes and the functions of conserved miRNAs. The expression levels of the two representative potential target mRNAs (TC189536 and DR054135) were inversely proportional with the expression levels of their corresponding miRNAs (pta-miR166 and pta-miR951, respectively). Computational prediction shows that most of the targets of the differentially expressed miRNAs in the male gametophyte of loblolly pine appear to be involved in stress/defense response, followed by metabolism, gene regulation, and signaling. However, target functions based on the psRNATarget program are not available for miR901, miR903, and miR1125, which are all up-regulated in germinated pollen. Interestingly, these three miRNAs are among the significantly up-regulated miRNAs in response to drought stress in rice (Zhou et al. 2010). Stress/defense response-related transcripts are also preferentially expressed in rice pollen, which, according to Wei et al. (2010), indicates that the ability to deal with abiotic and biotic stresses during pollen development may be essential to successful fertilization.
This study has identified several interesting conserved miRNAs that are expressed in the male gametophyte of loblolly pine. This study further demonstrates that expressions of many conserved miRNAs are different between mature and germinated loblolly pine pollen, with most of the miRNAs are up-regulated in germinated pollen. This indicates that the transition from mature (ungerminated pollen) to germinated pollen is modulated at the miRNA level. This study also allowed comparisons of the type and expression patterns of some conserved miRNA between the male gametophytes of loblolly pine with those of rice and Arabidopsis. Although some similarities and differences have been identified, it is premature to identify any specific miRNA at this point. Nevertheless, it is interesting that such differentiation may be achieved even with just the use of conserved miRNAs. Since it was not the aim of the current study, further studies are needed to verify the miRNA targets and the specific roles they play in phase transitions, particularly from ungerminated to germinated phase of the male gametophyte. However, loblolly pine (and gymnosperms in general) is currently not amenable to molecular genetic analysis because of its long generation time. Therefore, functional analysis has to be delayed until alternative approaches become available including perhaps, the use of direct pollen or protoplast transient transformation as a system for single-cell analysis which is emerging as the new frontier in genomic research.
References
Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC (2005) Cloning and characterization of microRNAs from moss. Plant J 43:837–848
Axtell MJ, Bartel DP (2005) Antiquity of microRNAs and their targets in land plants. Plant Cell 17:1658–1673
Borges F, Pereira PA, Slotkin RK, Martienssen RA, Becker JD (2011) MicroRNA activity in the Arabidopsis male gametophyte. J Exp Bot 62:1611–1620
Brewbaker JL, Kwack BH (1963) The essential role of calcium ion in pollen germination and pollen tube growth. Am J Bot 50:859–865
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Chambers C, Shuai B (2009) Profiling microRNA expression in Arabidopsis pollen using microRNA array and real-time PCR. BMC Plant Biol 9:87
Chen CC (2009) Small RNAs and their roles in plant development. Annu Rev Cell Biol 25:21–44
Chen Y, Chen T, Shen S, Zheng M, Guo Y, Lin J, Baluska F, Samai J (2006) Differential display proteomic analysis of Picea meyeri pollen germination and pollen tube growth after inhibition of actin polymerization by latrunculin B. Plant J 47:174–195
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159
Dai S, Chen T, Chong K, Xue Y, Liu S, Wang T (2007) Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol Cell Proteomics 6:207–230
Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ (2008) Conifers have a unique small RNA silencing signature. RNA 14:1508–1515
Fernando DD, Owens JN, von Aderkas P, Takaso T (1997) In vitro pollen tube growth and penetration of female gametophyte in Douglas fir (Pseudotsuga menziesii). Sex Plant Reprod 10:209–216
Fernando DD, Quinn CR, Brenner ED, Owens JN (2010) Male gametophyte development and evolution in extant gymnosperms. Int J Plant Dev Bio 4:47–63
Floyd SK, Bowman JL (2004) Gene regulation: ancient microRNA target sequences in plants. Nature 428:485–486
Fujioka T, Kaneko F, Kazama T, Suwabe K, Suzuki G, Makino A, Mae T, Endo M, Kawagishi-Kobayashi M, Watanabe M (2008) Identification of small RNAs in late developmental stage of rice anthers. Genes Genet Syst 83:281–284
Gifford EM, Foster AS (1989) Morphology and evolution of vascular plants. W. H. Freedman and Company, New York
Grant-Downton R (2010) Through a generation darkly: small RNAs in the gametophyte. Biochem Soc Trans 38:617–621
Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H (2009) MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10:643
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8:38–44
Le Trionnaire G, Twell D (2010) Small RNAs in angiosperm gametophytes: from epigenetics to gamete development. Gene Dev 24:1081–1085
Le Trionnaire G, Grant-Downton RT, Kourmpetli S, Dickinson HG, Twell D (2011) Small RNA activity and function in angiosperm gametophytes. J Exp Bot 62:1601–1610
Lelandais-Briere C, Sorin C, Declerck M, Benslimane A, Crespi M, Hartmann C (2010) Small RNA diversity in plants and its impact in development. Curr Genomics 11:14–23
Lu SF, Sun YH, Shi R, Clark C, Li LG, Chiang VL (2005) Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17:2186–2203
Lu SF, Sun YH, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51:1077–1098
Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38:S31–S36
Morin RD, Askay G, Dolgosheina D, Ebhardt HA, Magrinio V, Mardiso ER, Cenk Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18:571–584
Nystedt et al (2013) The Norway spruce genome sequence and conifer genome evolution. doi:10.1038/nature12211
Oh TJ, Wartell RM, Cairney J, Pullman GS (2008) Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytol 179:67–80
Peng H, Cun J, Ai TB, Tong YA, Zhang R, Zhao MM, Chen F, Wang SH (2012) MicroRNA profiles and their control of male gametophyte development in rice. Plant Mol Biol 80:85–102
Qiu D, Xiaoping P, Wilson IW, Li F, Liu M, Teng W, Zhang B (2009) High-throughput sequencing technology reveals that the taxoid elicit methyl jasmonate regulated microRNA expression in Chinese yew (Taxus chinensis). Gene 432:37–44
Singh H (1978) Embryology of gymnosperms. Gebruder Borntraegar, Berlin
Wan LC, Wang F, Guo X, Lu S, Qiu Z, Zhao Y, Zhang H, Lin J (2012a) Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol 12:146
Wan LC, Zhang H, Lu S, Zhang L, Qiu Z, Zhao Y, Zeng QY, Lin J (2012b) Transcriptome-wide identification and characterization of miRNAs from Pinus densata. BMC Biol 13:132
Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T (2010) Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11:338
Wei LQ, Yan LF, Wang T (2011) Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol 12:R53
Williams J (2012) Pollen tube growth rates and the diversification of flowering plant reproductive cycles. Int J Plant Sci 173:649–661
Yakovlev IA, Fossdal CG, Johnsen O (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytol 187:1154–1169
Zhan S, Lukens L (2010) Identification of novel miRNAs and miRNA dependent developmental shifts of gene expression in Arabidopsis thaliana. PLoS One 5:e10157
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33:W701–W704
Zhang BH, Pan XP, Cannon CH, Cobb GP, Anderson TA (2006a) Conservation and divergence of plant microRNA genes. Plant J 46:243–259
Zhang B, Pan X, Anderson TA (2006b) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang S, Zhou J, Han S, Yang W, Li W, Wei H, Li X, Qi L (2010) Four abiotic stress-induced miRNA families differentially regulated in the embryonic and non-embryonic callus tissues of Larix leptolepis. Biochem Biophs Res Commun 398:355–360
Zhang J, Zhang S, Han S, Wu T, Li X, Li W, Qi L (2012) Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236:647–657
Zhang J, Wu T, Li L, Han S, Li X, Zhang S, Qi L (2013) Dynamic expression of small RNA populations in larch (Larix leptolepis). Planta 237:89–101
Zhao C-Z, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ, Li M-, Li CS, Zhang BH, Wang XJ (2010) Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol 10:3–14
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61:4157–4168
Zhu Y, Zhao P, Wu X, Wang W, Scali M, Cresti M (2011) Proteomic identification of differentially expressed proteins in mature and germinated maize pollen. Acta Physiol Plant 33:1467–1474
Zou J, Song L, Zhang W, Wang Y, Ruan S, Wu W-H (2009) Comparative proteomic analysis of Arabidopsis mature pollen and germinated pollen. J Int Plant Biol 51:438–455
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Venkatesan Sundaresan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Quinn, C.R., Iriyama, R. & Fernando, D.D. Expression patterns of conserved microRNAs in the male gametophyte of loblolly pine (Pinus taeda). Plant Reprod 27, 69–78 (2014). https://doi.org/10.1007/s00497-014-0241-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-014-0241-3