Abstract
Gordon Syndrome (GS) is a rare familial hypertension syndrome with a characteristic hyperkalaemia which distinguishes it from other syndromic forms of hypertension that typically cause hypokalaemia. Patients with GS respond to aggressive salt-restriction or relatively small doses of thiazide diuretics, which suggests that activation of the thiazide-sensitive Na/Cl cotransporter (NCC) in the distal nephron is to blame. However, the mechanism has proved to be complex. In 2001, mutations in genes encoding two serine/threonine kinases, WNK1 and WNK4, were identified as causing GS. However, it took several years to appreciate that these kinases operated in a cascade with downstream serine/threonine kinases (SPAK and OSR1) actually phosphorylating and activating NCC and the closely related cotransporters NKCC1 and NKCC2. The hyperkalaemia in GS arises from an independent action of WNK1/WNK4 to reduce cell-surface expression of ROMK, the secretory K-channel in the collecting ducts. However, mutations in WNK1/4 are present in a small minority of GS families, and further genes have emerged (CUL3 and KLHL3) that code for Cullin-3 (a scaffold protein in an ubiquitin–E3 ligase) and an adaptor protein, Kelch3, respectively. These new players regulate the ubiquitination and proteasomal degradation of WNK kinases, thereby adding to the complex picture we now have of NCC regulation in the distal nephron.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first report of a patient with hypertension and striking hyperkalaemia appeared in the 1960s [1]. Richard Gordon reported a second case in 1964 [2], but soon realised that it was an inheritable phenotype tracking in several Australian families. Patients with this phenotype have normal renal function in terms of an intact glomerular filtration rate and usually a metabolic acidosis (often called type IV renal tubular acidosis). The hyperkalaemia itself can be severe (reaching 8–9 mmol/l) and sufficient to cause symptoms of periodic paralysis leading to the alternative name for the syndrome of familial hyperkalaemia and hypertension (FHHt); pseudohypoaldosteronism type 2 (PHA2) is another syndrome of hypertension characterised by hyperkalaemia. The hyperkalaemia is a useful discriminator because all other monogenic syndromes that increase blood pressure cause no change in plasma potassium (K) or obvious hypokalaemia [3].
Subsequently pedigrees with the Gordon Syndrome (GS) phenotype have been reported from France [4] and Israel [5]. The French pedigrees were notable because they were associated with a much milder phenotype. The implications of this milder phenotype would become better understood with the realisation that there are distinct phenotype–genotype correlations across GS families (see below).
Patients with GS usually have suppressed renin consistent with their salt-loaded state, but the aldosterone levels are typically low given their hyperkalaemia. Both conditions are reversed by dietary salt-restriction (20 mmol/day). In fact, either salt-restriction or low doses of thiazide diuretics are highly effective in normalising the hypertension and electrolyte disturbances of patients with GS [5, 6].
Early linkage studies and discovery of WNK1/4 mutations
The GS phenotype usually tracks through families as an autosomal dominant trait, although the most recent molecular work has indicated that some mutations causing a GS phenotype behave recessively (KLHL3, see below). The initial genetic studies with GS pedigrees were hampered by the small size of many of the families and hence their limited use in linkage studies to define candidate loci. However, it was clear at an early stage that there was linkage heterogeneity, and two loci were ultimately defined on chromosomes 1 (1q31-q42) and 17 (17p11-q21) [7, 8]. The breakthrough came during linkage studies on a large French pedigree that showed an intronic deletion in a third locus on chromosome 12 [9]. This deletion occurred in a gene called WNK1 that had been recently characterised as a gene encoding a serine/threonine kinase. Indeed, WNK1 was a member a small family of four WNK kinases (WNK1–4) with highly conserved features [10]. Another member of this family, WNK4, actually mapped to the chromosome 17 locus, so it was soon established that families linking to this locus carried WNK4 mutations. However, these mutations were exonic missense mutations that clustered in a unique amino acid sequence (EPEEPEADQH) dubbed the acidic motif. All of these mutations caused charge-changing substitutions in the motif, but they did not affect the kinase activity of WNK4. In fact, the function of the acidic motif remained a mystery until the recent discovery that the WNKs bind to Kelch-3 through the acidic motif [11, 12] (see below). In contrast, the intronic deletion in WNK1 appears to function by increasing WNK1 expression—at least in the circulating peripheral blood monocytes from GS patients with WNK1 mutations [9].
The function of WNK kinases in the kidney
Throughout the human kinome, kinase enzymes almost invariably use a canonical lysine residue to dock ATP. The singular exceptions are the WNK kinases, hence their acronym [‘With No lysine (K)’]. As a group the WNK kinases are highly conserved and phylogenetically old, appearing first in the Drosophila and Caenorhabditis elegans genomes (WNK1), with the other WNKs only appearing in higher vertebrates, such as the Fugu genome and upwards. Early clues to the function of the WNKs came from their expression in transporting epithelia [13, 14] and the clinical parallel of GS with another familial blood pressure and electrolyte disorder, Gitelman syndrome. Indeed, GS and Gitelman syndrome were found to be mirror images of each other, which suggested that the thiazide-sensitive NaCl cotransporter (NCC) was involved and probably activated. However, the coding sequence of NCC was not mutated in GS patients, and the GS loci identified by linkage did not map to the SLC12A3 gene encoding NCC (chromosome 16q3).
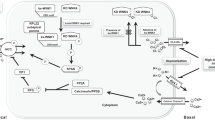
To address this puzzle, several groups looked at co-expression of NCC and WNK proteins using the oocyte expression system [15–17] and found that WNK4 suppressed total Na flux by a marked inhibition of NCC expression at the cell surface. The effect was reversed when WNK4 was expressed with typical GS mutations in its acidic motif. Further work has shown that WNK4 does this by blocking forward trafficking of NCC, which is instead diverted for early lysosomal degradation [18]. However, this is not the only—or probably even the major—regulatory effect of WNKs on NCC, because a second question remained: if the WNK proteins are kinases, what is their phosphorylation target? The WNKs do not phosphorylate NCC directly, and their kinase activity against prototypical substrates, such myelin basic protein and claudin-4, is very weak. Then in 2005, Dario Alessi’s group in Dundee identified two related proteins, SPAK (STE20/SPS1-related proline/alanine-rich kinase) and OSR1 (oxidative stress–responsive kinase 1), which were very actively phosphorylated by WNKs [19]. In fact, these closely related STE20 serine/threonine kinases are inactive until a single threonine residing in their catalytic loops (T233 in SPAK and T185 OSR1) are phosphorylated by WNKs. The phospho-forms of both kinases then in turn phosphorylate key threonine residues in the N-terminal region of NCC and in the related chloride-cation cotransporters NKCC1 and NKCC2 [20]. Phosphorylation of these residues increases transporter activity in association with insertion of the transporter into the apical surface membrane [21–23] (Fig. 1).
Different actions of the ‘With No lysine (K)’ kinases (WNK s) along the distal nephron. WNK1/4 phosphorylates the STE20/SPS1-related proline/alanine-rich kinase (SPAK) in the distal convoluted tubule (DCT), which in turn phosphorylates and activates the thiazide-sensitive Na/Cl cotransporter (NCC). This occurs in tandem with insertion of the phosphorylated NCC into the apical membrane. In the adjacent collecting duct (CD), WNK1/4 accelerates internalisation of the secretory K-channel in the CD (ROMK channel). ENaC Epithelial sodium channel
The importance of this mechanism in vivo can be seen when the key N-terminal threonine residue in NCC (T60) is inactivated by a loss-of-function mutation (T60M). This is a common disease allele amongst South-East Asians with Gitelman syndrome, and the low apical expression of NCC leads to reduced recovery of the protein from urinary exosomes [24]. A final twist to this story has come with a report that the activation of NCC by WNK1 is independent of WNK4 and is, in fact, actually antagonised by the latter [25]. This suggests that in vivo the WNKs (WNK1, WNK3 and WNK4) form homo- and hetero-oligomers through their coil–coil domains to regulate NCC.
The origin of the hyperkalaemia in GS
The mechanism for the salt-dependent hypertension can be explained by NCC activation, but what is the origin of the hallmark hyperkalaemia? It too could be explained in part by NCC activation. The effect of NCC activation is to extract more NaCl from the distal convoluted tubule (DCT), which leads to reduced electrogenic recovery of Na+ in the collecting ducts (CD) (Fig. 1) and thereby a reduction in the lumen-negative potential required for K secretion through the secretory K-channel in the CD (ROMK). However, several groups have shown that the WNKs are able to directly affect ROMK expression by affecting its trafficking from the cell surface [26–28]. Specifically, the WNKs accelerate the endocytosis of ROMK through a clathrin-coated pit pathway (Fig. 1). The GS mutations in WNK4 do not block this behaviour but actually behave as gain-of-function mutations suppressing ROMK expression even further [17]. It therefore seems likely that the hyperkalaemia arises from a combination of a reduced potential across individual ROMK channels as well as a reduction in the numbers of channels expressed in the CD cells in GS. Nevertheless, the importance of altered endocytosis has been questioned, but in a recent WNK1 mouse model of GS the expression of the ROMK was clearly downregulated in the CD of these mice in vivo [29].
WNKs are not the whole story
The original linkage work in GS pedigrees implied that mutations in at least four genes could cause a GS phenotype. To explore this, Rick Lifton’s research group undertook a whole exome approach in 2012, which succeeded in identifying two further genes, CUL3 and KLHL3 [30]. A systematic screening of their pedigree collection showed that mutations in these genes actually accounted for the overwhelming majority of disease-causing mutations, with just seven of 56 GS families being explained by a mutation in either WNK1 or WNK4. In support of this finding, mutations in Kelch-3, encoded by the KLHL3 gene [31], have been reported in 16 of 45 French GS pedigrees, and mutations in CUL3 and KLHL3 have been detected in a combined UK and Australian collection of 11 GS pedigrees without a previous molecular diagnosis [32].
The proteins encoded by CUL3 and KLHL3 are involved the endosomal degradation of WNK4 and probably also for the WNK proteins as a class. Cullin-3 is actually the hydrophobic scaffold protein for a ubiquitin–E3 ligase that tags proteins, in this case WNKs, for degradation through the proteasome (Fig. 2). The substrate specificity for the E3 ligase is dictated by the substrate adaptor protein for the Cullin-RING (really interesting new gene) complex and that for the WNKs is provided by Kelch-3. The functional implications for the CUL3 and KLHL3 mutations found in GS fit well with the known structural biology of the Cullin-RING-ligase–Kelch interaction. Hence, all of the CUL3 mutations affect the splicing of exon 9 of Cullin3, as is shown by the truncated transcripts expressed in patients with these mutations [32]. It is thought that the 57-residue exon 9 peptide normally interacts directly with Kelch-3 and that therefore the CUL3 mutations will block the Cullin3 interaction with Kelch-3. The Kelch domains themselves (there are 6 of them) in Kelch-3 form a propeller-like structure, and the dominant mutations lie either close to the hub of the propeller or at sites thought to be important for binding the substrate protein (WNKs in this case) [30, 31]. The implication of this work is that the levels of WNKs should be increased in cells expressing these Cullin3 or Kelch-3 mutations. This upregulation has been confirmed in a recent Kelch-3 mouse model for a KLHL3 GS mutation, with the tissues of these animals showing widespread upregulation of WNK4 [33].
The Cullin3–E3 ligase complex. The complex binds WNKs through the adaptor protein Kelch-3 (encoded by KLHL3) and the WNK acidic motif (EPEEPEADQH). The ubiquitinated WNKs are then degraded through the proteasome. The Gordon Syndrome (GS) mutations CUL3 and KLHL3 block this pathway, causing the WNKs to accumulate in the cell. Ub–E2 ubiquitin–E2 ligase
Discovery of the CUL3 and KLHL3 mutations has led to the emergence of clear phenotype–genotype correlations in GS [30, 32] (Fig. 3). Mutations in CUL3 invariably cause onset of the GS phenotype in very early childhood and the most profound electrolyte disturbances. It therefore cannot come as a surprise that Richard Gordon’s very severe 1964 index case carries a CUL3 mutation [32]. The age of onset and severity then diminishes for the other genes in the order: KLHL3 > WNK4 > WNK1. In fact, the WNK1 families typically have normal blood pressure into late adulthood [4, 34].
Genotype–phenotype relations for the various genes mutated in GS. CUL3 carries the most severe phenotype expressed in terms of age of diagnosis, degree of hyperkalaemia and/or metabolic acidosis. AD Autosomal dominant, AR autosomal recessive. Figure is redrawn from Boyden et al. [30] with permission
A vascular component to the hypertension in GS?
It has been assumed that the hypertension seen in GS is entirely explained by salt-retention through NCC activation. However, this may not always be the case—at least for those cases caused by CUL3 mutations. It was reported over 20 years ago that dominant negative mutations in PPARG, the target for thiazolidinedione antidiabetic drugs, had a striking phenotype of severe insulin resistance, diabetes and marked hypertension [35]. The molecular basis for this was then unclear. However, recent work has shown that these PPARG mutations cause vasoconstriction by an intriguing mechanism [36]. The contractile tone of vascular smooth muscle is controlled by the phophosphorylation state of myosin-light chain that is determined by RhoA-kinase. This is in turn regulated by RhoA, a molecule that is subject to degradation through ubiquitinylation and endosomal breakdown in an analogous fashion to the WNKs. RhoA even uses a Cullin3–E3 RING ligase, although it partners a different adaptor protein, RhoBTB1. Hence, it would be expected that GS subjects carrying typical CUL3 exon 9 deletions would also show defective RhoA ubiquitinylation and that its accumulation would lead to increased vascular smooth muscle tone. This mechanism would provide an elegant explanation for the more severe blood pressure phenotype seen with CUL3 versus KLHL3 mutations.
GS-causing mutations in the general hypertensive population?
Pedigrees with GS are uncommon, with fewer than 150 reported in the literature world-wide. However, an obvious question is whether GS-causing mutations are under-recognised in the wider hypertensive population. Louis-Dit-Picard et al. have directly sequenced the coding region of KLHL3 in 1,232 hypertensive subjects from the HYPERGENE cohort and found just a single missense mutation in a subject with a plasma K level of 5.1 mmol/l [31]; no such missense mutation was present in 800 normotensive control subjects. The same research group also looked for an association between blood pressure and almost 800 single nucleotide polymorphisms (SNPs) across the KLHL3 locus in another cohort (International Consortium on Blood Pressure) of 69,000 subjects—but found none. So it appears sporadic GS-causing KLHL3 mutations are rare in European subjects and that the gene itself does not influence blood pressure in the general population. This has also been the experience with WNK1 and WNK4, where a large meta-analysis of genome-wide association study cohorts have failed to show a significant association of either gene locus with hypertension in the general population. However, a significant association has been found with common SNP variants encoding SPAK (also known as STK39) that has been replicated across four large cohorts [37].
The future
It is now clear that GS can be caused by at least four genes that modulate a novel WNK/OSR1/SPAK/NCC regulatory pathway in the distal nephron. Understanding the molecular mechanisms involved has revealed the complexity with which NCC is regulated. NCC can no longer be seen as a static target for thiazide drugs, but rather as a highly dynamic cotransporter whose phosphorylation state and expression at the cell surface are tightly controlled. There is also substantial evidence that WNKs are important outside of the kidney with impacts on bone, central nervous system and the vasculature [38, 39]. Yet, the GS story is still incomplete because GS pedigrees remain that do not have mutations in CUL3, KLHL3 or the genes encoding WNK1 or WNK4. The roll-out of whole genome sequencing to these families may identity further GS genes. There is also the prospect that in some families (with CUL3 mutations) the phenotype has a vascular dimension that needs further exploration. Finally, the WNK/ OSR1/SPAK phosphorylation cascade is an attractive drugable target, thereby raising the prospect of a new class of diuretic drugs that block the docking of WNKs and SPAK. In fact, a Japanese group has recently used a high-throughput drug screen to identify two potential lead compounds targeting the peptide motif (RFQV) used to dock WNK with SPAK [38, 40]. So despite the extreme rarity of GS, the unfolding its molecular basis seems set to have a huge translational impact both inside and outside the kidney.
References
Paver W, Pauline G (1964) Hypertension and hyperpotassaemia without renal disease in a young male. Med J Aust 2:305–306
Gordon RD, Geddes RA, Pawsey CG, O’Halloran MW (1970) Hypertension and severe hyperkalaemia associated with suppression of renin and aldosterone and completely reversed by dietary sodium restriction. Australas Ann Med 19:287–294
Toka HR, Koshy JM, Hariri A (2013) The molecular basis of blood pressure variation. Pediatr Nephrol 28:387–399
Achard JM, Disse-Nicodeme S, Fiquet-Kempf B, Jeunemaitre X (2001) Phenotypic and genetic heterogeneity of familial hyperkalaemic hypertension (Gordon syndrome). Clin Exp Pharmacol Physiol 28:1048–1052
Mayan H, Vered I, Mouallem M, Tzadok-Witkon M, Pauzner R, Farfel Z (2002) Pseudohypoaldosteronism type II: marked sensitivity to thiazides, hypercalciuria, normomagnesemia, and low bone mineral density. J Clin Endocrinol Metab 87:3248–3254
Gordon RD, Hodsman GP (1986) The syndrome of hypertension and hyperkalaemia without renal failure: long term correction by thiazide diuretic. Scott Med J 31:43–44
Mansfield TA, Simon DB, Farfel Z, Bia M, Tucci JR, Lebel M, Gutkin M, Vialettes B, Christofilis MA, Kauppinen Makelin R, Mayan H, Risch N, Lifton RP (1997) Multilocus linkage of familial hyperkalaemia and hypertension, pseudohypoaldosteronism type II, to chromosomes 1q31–42 and 17p11–q21. Nat Genet 16:202–205
O’Shaughnessy KM, Fu B, Johnson A, Gordon RD (1998) Linkage of Gordon’s syndrome to the long arm of chromosome 17 in a region recently linked to familial essential hypertension. J Hum Hypertens 12:675–678
Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP (2001) Human hypertension caused by mutations in WNK kinases. Science 293:1107–1112
Verissimo F, Jordan P (2001) WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 20:5562–5569
Uchida S, Sohara E, Rai T, Sasaki S (2014) Regulation of with-no-lysine kinase signaling by Kelch-like proteins. Biol Cell 106:45–56
Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T (2013) The CUL3-KLHL3 E3 ligase complex mutated in Gordon’s hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease-causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 45:111–122
Kahle KT, Gimenez I, Hassan H, Wilson FH, Wong RD, Forbush B, Aronson PS, Lifton RP (2004) WNK4 regulates apical and basolateral Cl- flux in extrarenal epithelia. Proc Natl Acad Sci USA 101:2064–2069
Choate KA, Kahle KT, Wilson FH, Nelson-Williams C, Lifton RP (2003) WNK1, a kinase mutated in inherited hypertension with hyperkalemia, localizes to diverse Cl–transporting epithelia. Proc Natl Acad Sci USA 100:663–668
Yang CL, Angell J, Mitchell R, Ellison DH (2003) WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest 111:1039–1045
Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP (2003) Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci USA 100:680–684
Golbang AP, Murthy M, Hamad A, Liu CH, Cope G, Van’t Hoff W, Cuthbert A, O’Shaughnessy KM (2005) A new kindred with pseudohypoaldosteronism type II and a novel mutation (564D>H) in the acidic motif of the WNK4 gene. Hypertension 46:295–300
Zhou B, Zhuang J, Gu D, Wang H, Cebotaru L, Guggino WB, Cai H (2010) WNK4 enhances the degradation of NCC through a Sortilin-mediated lysosomal pathway. J Am Soc Nephrol 21:82–92
Vitari AC, Deak M, Morrice N, Alessi DR (2005) The WNK1 and WNK4 protein kinases that are mutated in Gordon’s hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391:17–24
Alessi DR, Zhang J, Khanna A, Hochdorfer T, Shang Y, Kahle KT (2014) The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci Signal 7:re3
Rosenbaek LL, Kortenoeven MLA, Aroankins TS, Fenton RA (2014) Phosphorylation decreases ubiquitylation of the Thiazide-sensitive cotransporter NCC and subsequent clathrin-mediated endocytosis. J Biol Chem 289:13347–13361
Glover M, Mercier Zuber A, Figg N, O’Shaughnessy KM (2010) The activity of the thiazide-sensitive Na(+)-Cl(-) cotransporter is regulated by protein phosphatase PP4. Can J Physiol Pharmacol 88:986–995
Pacheco-Alvarez D, Cristobal PS, Meade P, Moreno E, Vazquez N, Munoz E, Diaz A, Juarez ME, Gimenez I, Gamba G (2006) The Na+:Cl- cotransporter is activated and phosphorylated at the amino-terminal domain upon intracellular chloride depletion. J Biol Chem 281:28755–28763
Yang S-S, Fang Y-W, Tseng M-H, Chu P-Y, Yu I-S, Wu H-C, Lin S-W, Chau T, Uchida S, Sasaki S, Lin Y-F, Sytwu H-K, Lin S-H (2013) Phosphorylation regulates NCC stability and transporter activity in vivo. J Am Soc Nephrol 24:1587–1597
Chavez-Canales M, Zhang C, Soukaseum C, Moreno E, Pacheco-Alvarez D, Vidal-Petiot E, Castaneda-Bueno M, Vazquez N, Rojas-Vega L, Meermeier NP, Rogers S, Jeunemaitre X, Yang CL, Ellison DH, Gamba G, Hadchouel J (2014) WNK-SPAK-NCC cascade revisited: WNK1 stimulates the activity of the Na-Cl cotransporter via SPAK, an effect antagonized by WNK4. Hypertension. doi:10.1161/HYPERTENSIONAHA.114.04036
Cope G, Golbang A, Murthy M, Hamad A, Liu CH, Hoff WV, Cuthbert A, O’Shaughnessy KM (2006) WNK1 affects surface expression of the ROMK potassium channel independent of WNK4. J Am Soc Nephrol 17:1867–1874
Kahle KT, Wilson FH, Leng Q, Lalioti MD, O’Connell AD, Dong K, Rapson AK, MacGregor GG, Giebisch G, Hebert SC, Lifton RP (2003) WNK4 regulates the balance between renal NaCl reabsorption and K+ secretion. Nat Genet 35:372–376
Lazrak A, Liu Z, Huang CL (2006) Antagonistic regulation of ROMK by long and kidney-specific WNK1 isoforms. Proc Natl Acad Sci USA 103:1615–1620
Vidal-Petiot E, Elvira-Matelot E, Mutig K, Soukaseum C, Baudrie V, Wu S, Cheval L, Huc E, Cambillau M, Bachmann S, Doucet A, Jeunemaitre X, Hadchouel J (2013) WNK1-related familial hyperkalemic hypertension results from an increased expression of L-WNK1 specifically in the distal nephron. Proc Natl Acad Sci USA 110:14366–14371
Boyden LM, Choi M, Choate KA, Nelson-Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G, Lebel M, Gordon RD, Semmekrot BA, Poujol A, Valimaki MJ, De Ferrari ME, Sanjad SA, Gutkin M, Karet FE, Tucci JR, Stockigt JR, Keppler-Noreuil KM, Porter CC, Anand SK, Whiteford ML, Davis ID, Dewar SB, Bettinelli A, Fadrowski JJ, Belsha CW, Hunley TE, Nelson RD, Trachtman H, Cole TR, Pinsk M, Bockenhauer D, Shenoy M, Vaidyanathan P, Foreman JW, Rasoulpour M, Thameem F, Al-Shahrouri HZ, Radhakrishnan J, Gharavi AG, Goilav B, Lifton RP (2012) Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482:98–102
Louis-Dit-Picard H, Barc J, Trujillano D, Miserey-Lenkei S, Bouatia-Naji N, Pylypenko O, Beaurain G, Bonnefond A, Sand O, Simian C, Vidal-Petiot E, Soukaseum C, Mandet C, Broux F, Chabre O, Delahousse M, Esnault V, Fiquet B, Houillier P, Bagnis CI, Koenig J, Konrad M, Landais P, Mourani C, Niaudet P, Probst V, Thauvin C, Unwin RJ, Soroka SD, Ehret G, Ossowski S, Caulfield M, Bruneval P, Estivill X, Froguel P, Hadchouel J, Schott JJ, Jeunemaitre X (2012) KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44:456–460
Glover M, Ware JS, Henry A, Wolley M, Walsh R, Wain LV, Xu S, Van’t Hoff WG, Tobin MD, Hall IP, Cook S, Gordon RD, Stowasser M, O’Shaughnessy KM (2014) Detection of mutations in KLHL3 and CUL3 in families with FHHt (familial hyperkalaemic hypertension or Gordon’s syndrome). Clin Sci (Lond) 126:721–726
Susa K, Sohara E, Rai T, Zeniya M, Mori Y, Mori T, Chiga M, Nomura N, Nishida H, Takahashi D, Isobe K, Inoue Y, Takeishi K, Takeda N, Sasaki S, Uchida S (2014) Impaired degradation of WNK1 and WNK4 kinases causes PHAII in mutant KLHL3 knock-in mice. Hum Mol Genet. doi:10.1093/hmg/ddu217
Disse-Nicodeme S, Desitter I, Fiquet-Kempf B, Houot AM, Stern N, Delahousse M, Potier J, Ader JL, Jeunemaitre X (2001) Genetic heterogeneity of familial hyperkalaemic hypertension. J Hypertens 19:1957–1964
Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O’Rahilly S (1999) Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 402:880–883
Pelham CJ, Ketsawatsomkron P, Groh S, Grobe JL, de Lange WJ, Ibeawuchi SR, Keen HL, Weatherford ET, Faraci FM, Sigmund CD (2012) Cullin-3 regulates vascular smooth muscle function and arterial blood pressure via PPARgamma and RhoA/Rho-kinase. Cell Metab 16:462–472
Wang Y, O’Connell JR, McArdle PF, Wade JB, Dorff SE, Shah SJ, Shi X, Pan L, Rampersaud E, Shen H, Kim JD, Subramanya AR, Steinle NI, Parsa A, Ober CC, Welling PA, Chakravarti A, Weder AB, Cooper RS, Mitchell BD, Shuldiner AR, Chang YP (2009) Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci USA 106:226–231
Alessi DR, Zhang J, Khanna A, Hochdörfer T, Shang Y, Kahle KT (2014) The WNK-SPAK/OSR1 pathway: master regulator of cation-chloride cotransporters. Sci Signal 7(334):re3
Siew K, O’Shaughnessy KM (2013) Extrarenal roles of the with-no-lysine[K] kinases (WNKs). Clin Exp Pharmacol Physiol 40:885–894
Mori T, Kikuchi E, Watanabe Y, Fujii S, Ishigami-Yuasa M, Kagechika H, Sohara E, Rai T, Sasaki S, Uchida S (2013) Chemical library screening for WNK signalling inhibitors using fluorescence correlation spectroscopy. Biochem J 455:339–345
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
O’Shaughnessy, K.M. Gordon Syndrome: a continuing story. Pediatr Nephrol 30, 1903–1908 (2015). https://doi.org/10.1007/s00467-014-2956-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00467-014-2956-7