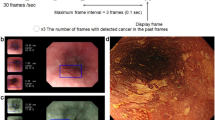
Abstract
Background
Diagnosis of early gastric cancer (EGC) under narrow band imaging endoscopy (NBI) is dependent on expertise and skills. We aimed to elucidate whether artificial intelligence (AI) could diagnose EGC under NBI and evaluate the diagnostic assistance of the AI system.
Methods
In this retrospective diagnostic study, 21,785 NBI images and 20 videos from five centers were divided into a training dataset (13,151 images, 810 patients), an internal validation dataset (7057 images, 283 patients), four external validation datasets (1577 images, 147 patients), and a video validation dataset (20 videos, 20 patients). All the images were labeled manually and used to train an AI system using You look only once v3 (YOLOv3). Next, the diagnostic performance of the AI system and endoscopists were compared and the diagnostic assistance of the AI system was assessed. The accuracy, sensitivity, specificity, and AUC were primary outcomes.
Results
The AI system diagnosed EGCs on validation datasets with AUCs of 0.888–0.951 and diagnosed all the EGCs (100.0%) in video dataset. The AI system achieved better diagnostic performance (accuracy, 93.2%, 95% CI, 90.0–94.9%) than senior (85.9%, 95% CI, 84.2–87.4%) and junior (79.5%, 95% CI, 77.8–81.0%) endoscopists. The AI system significantly enhanced the performance of endoscopists in senior (89.4%, 95% CI, 87.9–90.7%) and junior (84.9%, 95% CI, 83.4–86.3%) endoscopists.
Conclusion
The NBI AI system outperformed the endoscopists and exerted potential assistant impact in EGC identification. Prospective validations are needed to evaluate the clinical reinforce of the system in real clinical practice.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Gastric cancer (GC) is the fifth most common malignancy and ranks as the fourth leading cause of cancer-related deaths worldwide [1]. Typically, most GC patients are diagnosed at an advanced stage with a 5-year survival rate of < 20% [2]. If GC lesions are detected and diagnosed at an early stage, these lesions can be curatively resected with a 5-year survival rate of > 95% [2]. However, early gastric cancer (EGC) only exhibits subtle changes in the mucosa and can often be overlooked by the standard modality of white light imaging endoscopy (WLI). Studies have reported that miss rates for GC and precancerous lesions range from 18.3 to 40.0% in East Asian countries, making a relatively low diagnostic rate of EGC [3,4,5].
Narrow band imaging (NBI) is reported to show better performance for EGC diagnosis than WLI. A recent meta-analysis revealed that the sensitivity and specificity of NBI in EGC diagnosis were 86.0% and 96.0%, while the sensitivity and specificity of WLI were only 57.0% and 79.0% [6]. However, substantial expertise and skills are required to achieve good performance in using NBI in EGC diagnosis. It is reported that the accuracy of NBI in EGC diagnosis is relatively low in less experienced endoscopists [7,8,9]. Moreover, the diagnostic discrepancy of NBI is wildly existed, even among experienced endoscopists [10]. Therefore, it is quite valuable to develop practical tools to assist endoscopists in EGC diagnosis under NBI.
Artificial intelligence (AI) has wildly been used in assisting esophagogastroduodenoscopy [11,12,13]. Our previous studies have established AI systems based on deep convolutional neural networks (DCNN) to detect EGC and predict the invasion depth of GC in real-time [14, 15]. Three previous studies have also developed AI models in identifying EGC under NBI with a sensitivity of 79.2–98.0% and a specificity of 74.5–100.0% [16,17,18]. However, several inherent limitations, including relatively small sample size, unsatisfactory generalization, and inability to diagnose a lesion in real-time, have restrained the clinical applicability of these models. Moreover, real-time AI systems are often expensive and complicated to be deployed in endoscopic centers (especially in rural areas).
In this study, we aimed to train and validate a generalized AI system, capable of diagnosing EGC under NBI in real-time. We also compared the performance of the AI system and endoscopists and evaluated the diagnostic assistance of the AI system. Finally, we developed an open-access AI website with multi-device compatibility to broaden the applicable scenarios.
Methods
Patients and study design
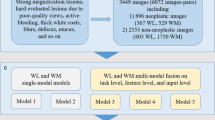
This retrospective, multicenter, diagnostic study was conducted based on the Helsinki declaration and the protocol was reviewed and approved by the Medical Ethics Committee of Nanjing University Medical School Affiliated Drum Tower Hospital (approval no. 2020–026-01). Due to the datasets being retrospectively established from deidentified patients, informed consent was not required. We developed and validated the DCNN system with datasets from five institutions in China: Nanjing University Medical School Affiliated Drum Tower Hospital (NJDTH), Wuxi People’s Hospital (WXPH), Taizhou People’s Hospital (TZPH), Nanjing Gaochun People’s Hospital (GCPH), and Changzhou Second People’s Hospital (CZPH). The study included a total of 1649 patients who underwent endoscopic submucosal dissection (ESD) following associated guidelines between January 2016 and February 2020 (Fig. 1). The inclusion criteria were as follows: a histological diagnosis of EGC; ESD treatment; and endoscopic examination before ESD. The exclusion criteria for patients were as follows: history of chemotherapy or radiation due to gastric cancer, gastric stump cancer, gastric lesions adjacent to the ulcer or ulcer scar, and multiple synchronous gastric cancerous lesions. The exclusion criteria for images were as follows: WLI, dye-stained imaging, ESD operation process, and poor quality (including less insufflation of air, halation, defocus, blurs, bubbles, sliding, fuzzy, and bleeding).
Preparation of training and validation datasets
After exclusion, a total of 20,208 endoscopic images from 1093 patients from NJDTH were used to train and internal validate the DCNN model (Fig. 1). The training and validation datasets were divided by temporal sequence to guarantee the independence of the datasets. In detail, all the datasets used in this study were as follows:
-
(1)
The training dataset: 13,151 images of 810 patients from NJDTH between January 2016 and October 2018 (among these images, 11,852 contained cancerous lesions).
-
(2)
The internal validation dataset: 7057 images of 283 patients from NJDTH between November 2018 and January 2019 (among these images, 5925 contained cancerous lesions).
-
(3)
The external validation datasets: WXPH, 645 images of 27 patients between June 2019 and December 2019 (311 images contained malignant lesions); TZPH, 114 images of 50 patients between June 2019 and December 2019 (57 images had malignant lesions); GCPH, 244 images of 51 patients between June 2019 and December 2019 (122 images included cancerous lesions); and CZPH: 574 images of 19 patients between June 2019 and December 2019 (287 images had cancerous lesions).
-
(4)
The video dataset included 20 videos of 20 consecutive patients from NJDTH between January 2020 and February 2020.
-
(5)
The testing dataset that included 300 cancerous images and 300 control images (no malignant lesions in the images) was established by randomly selecting from the internal validation dataset to evaluate the diagnostic performance and assistance of the DCNN system. The control images comprised chronic non-atrophic gastritis, chronic atrophic gastritis, and erosion, with specifically description in the Supplements (Table S1). All images and videos were obtained using Olympus endoscopes (GIF-H260Z and GIF-H290Z; Olympus Medical Systems, Tokyo, Japan) with video processors (EVIS LUCERA CV260/CLV260SL, EVIS LUCERA ELITE CV290/CLV290SL, Olympus Medical Systems). The classification and annotation strategies were as described previously [14].
Development of deep convolutional neural network system
We used You look only once v3 (YOLOv3) to develop the DCNN model to diagnose EGC under NBI due to the extremely fast detection and multi-scale predictions (Figure S1). Each image was normalized to 416*416 to input into the network. The lesions would be detected in three scales at the 79th layer of the network, and the feature map was obtained via 32 times subsampling. Considering the input was 416 * 416, the characteristic graph was 13 * 13 here. Due to the high-frequencies subsampling, the receptive field of the feature map was on a relatively large scale, which was suitable for detecting lesions of relatively large size in the image. To realize a close-grained detection, the feature map of the 79th layer was unsampled again. Then, a concatenation operation was performed with the feature map of the 61st layer to obtain the closer-grained feature map of the 91st layer. After several convolution layers, the feature map of the 91st layer was also obtained, which was 16 times lower sampled than the input image. It had a medium-scale receptive field and was suitable for detecting medium-scale lesions. Finally, the feature map of the 91st layer was unsampled again and concatenated with the feature map of the 36th layer. The final obtained feature map was 8 times lower sampled than the input image. It had the smallest receptive field and was suitable for detecting lesions of small size.
Measurements of diagnostic performance
We evaluated the performance of the DCNN system as follows:
-
(1)
Firstly, the diagnostic performance of the DCNN system was evaluated using the internal validation dataset.
-
(2)
Secondly, the generalization of the DCNN system was assessed using the four external validation datasets from WXPH, TZPH, GCPH, and CZPH. The robustness of the DCNN system was determined with subgroup analysis according to the invasion depth of lesions (intraepithelial lesions, intramucosal lesions, and submucosal lesions) on the internal validation dataset.
-
(3)
Thirdly, the performance of the DCNN system and endoscopists was compared using the testing dataset. Seven endoscopists from five institutions were divided into two groups based on the level of expertise: 3 seniors (minimum of 10-year experience with 10,000 EGD examinations) and 4 juniors (2-year experience with 2,000 EGD examinations). These endoscopists were not engaged in the annotation of the image datasets, and were unknown to the clinical characteristics, and pathological results of all the included patients. The testing images were all mixed in scrambled order and assessed by the endoscopists.
-
(4)
Fourthly, the testing images were scrambled and evaluated by the DCNN system and endoscopists with the assistance of the DCNN system after 2 weeks of washout to assess the assistance of the DCNN system.
-
(5)
Finally, the performance of our DCNN system was tested using the video dataset.
Main outcomes
The main outcomes included the accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). For per-image analysis, the optimal cut-off value is determined based on the Youden index. When the DCNN system annotated a suspicious lesion with a confidence value more than the cut-off value, it was regarded as a positive prediction. The confidence value contained two parts, the Intersection-over-Union (IoU), which is defined as:
and the probability of classification [Pr(object)]. The total confidence value was defined as: Confidence value = Pr(object) × IoU. For per-patient analysis, all the images of per patient were processed with the DCNN system and the proportion of positive predictions was used to diagnose the cancer patients based on the optimal Youden index.
Statistical analysis
For descriptive statistics, continuous variables were presented as means and standard deviations (SD) or medians and interquartile ranges, and categorical variables were expressed as frequencies and percentages. The area under ROC (AUC) was used to assess the diagnostic performance of the DCNN system. A two-sided McNemar test was used to compare the differences of accuracy, sensitivity, specificity. Generalized score statistics was used to compare the differences of PPV and NPV [19]. Inter-observer agreements were measured using Cohen’s kappa coefficient. All the statistical analyses were performed using R software (Version 4.0.5, The R Foundation, https://www.r-project.org) and R studio (Version 1.4.1106, RStudio, PBC, https://www.rstudio.com).
Results
Diagnostic performance of the DCNN model
The clinical baseline characteristic of the enrolled patients is shown in Table 1 and specifically described in Supplements. On the internal validation dataset, the AUC, sensitivity, and specificity of the DCNN model were 0.947 (95% CI, 0.939–0.956), 98.0% (95% CI, 97.6–98.3%) and 85.2% (95% CI, 83.0–87.1%), respectively (Fig. 2a, Table 2). The PPV and NPV of the DCNN model were 97.2% (95% CI, 96.7–97.6%) and 88.9% (95% CI, 86.9–90.7%) on the internal validation dataset (Table 2). On the external validation datasets, the AUC, sensitivity, and specificity of the DCNN model were 0.888–0.951, 87.7–96.7%, and 81.1–91.3%, respectively (Fig. 2a, Table 2). The PPV and NPV of the DCNN model were 83.7%–91.0% and 87.3–96.1% on the external validation datasets (Table 2). The visualized results showed that the predictive box was consistent with the pathological results (Fig. 3a) and annotations by the experts (Fig. 3b). The performance of the DCNN model was then evaluated in subgroups of the internal validation dataset based on the invasion depth. The DCNN model showed a robust performance in different subgroups with the AUC, sensitivity, and specificity of 0.915–0.955, 97.4–98.1%, and 74.7–86.9% (Fig. 2b, Table S2). Considering the imbalanced sample distribution in the internal validation dataset, we further investigated the performance of the DCNN model in per-patient level. The data showed that the DCNN model also exhibited a stable performance in per-patient level with an AUC of 0.975, a sensitivity of 100.0% and a specificity of 96.0% (Fig. 2c, Table S3). We also evaluated the diagnostic performance of the DCNN system in EGD videos collected between January 2020 and February 2020. The DCNN system diagnosed all the lesions in all the 20 (100.0%) consecutive EGD videos (Video 1), demonstrating a robust performance of the DCNN system.
Receiver operating characteristic curves (ROC) illustrating the performance of the DCNN system for EGC diagnosis. a ROC presenting the diagnostic ability of the DCNN system to diagnose EGC in NJDTH, WXPH, TZPH, and CZPH validation datasets in per-image analysis. b ROC showing the diagnostic ability of the DCNN model system to diagnose EGC in subgroups of intraepithelial lesions, intramucosal lesions, and submucosal lesions in per-image analysis. c ROC presenting the diagnostic ability of the DCNN system to diagnose EGC in NJDTH validation datasets in per-patient analysis. DCNN, Deep convolutional neural network; EGC, Early gastric cancer; NJDTH, Nanjing University Medical School Affiliated Drum Tower Hospital; WXPH, Wuxi People’s Hospital; TZPH, Taizhou People’s Hospital; GCPH, Nanjing Gaochun People’s Hospital; CZPH, Changzhou Second People’s Hospital
Comparison between the DCNN system and endoscopists
We then compared the diagnostic performance of the DCNN system and endoscopists. On the testing dataset, the accuracy (93.2%, 95% CI, 90.9–94.9%), sensitivity (99.0%, 95% CI, 97.1–99.7%) and NPV (98.9%, 95% CI, 96.7–99.7%) of the DCNN system were significantly superior to the accuracy (85.9%, 95% CI, 84.2–87.4%, P < 0.001), sensitivity (83.2%, 95% CI, 80.6–85.5%, P < 0.001), and NPV (84.1%, 95% CI, 81.6–86.3%, P < 0.001) of the senior endoscopists (Table 3). Similarly, the accuracy, sensitivity, PPV, and NPV of the DCNN system were statistically higher than that of the junior endoscopists (P < 0.001) and were shown specifically in Table 3.
Thereafter, we evaluated the assistance of the DCNN system. With the assistance, the diagnostic accuracy (85.9%, 95% CI, 84.2–87.4% versus 89.4%, 95% CI, 87.9–90.7%, P < 0.001), sensitivity (83.2%, 95% CI, 80.6–85.5% versus 90.7%, 95% CI, 88.6–92.4%, P < 0.001), PPV (87.9%, 95% CI, 85.5–89.9% versus 88.4%, 95% CI, 86.2–90.3%, P = 0.034), and NPV (84.1%, 95% CI, 81.6–86.3% versus 90.4%, 95% CI, 88.3–92.2%, P < 0.001) of the seniors were significantly improved (Fig. 4a, Table 3). Consistently, the diagnostic accuracy (79.5%, 95% CI, 77.8–81.0% versus 84.9%, 95% CI, 83.4–86.3%, P < 0.001), sensitivity (72.3%, 95% CI, 69.7–74.8% versus 83.8%, 95% CI, 81.6–85.7%, P < 0.001), PPV (84.4%, 95% CI, 82.0–86.4% versus 85.8%, 95% CI, 83.6–87.6%, P < 0.001), and NPV (75.8%, 95% CI, 73.4–78.0% versus 84.1%, 95% CI, 82.0–86.1%, P < 0.001) of the juniors were also enhanced statistically (Fig. 4b, Table 3). Notably, the specificity of the seniors (88.6% versus 88.1%) and juniors (86.6% versus 86.1%) was decreased marginally (Fig. 4b, Table 3).
Comparison of the performance of endoscopists with or without the assistance of the DCNN system. a The diagnostic performance of senior endoscopists with or without the aid of the DCNN system. b The diagnostic performance of junior endoscopists with or without the assistance of the DCNN system. c The inter-observer agreement of endoscopists with or without aid of the DCNN system. d The workflow for the potential applicable scenarios of the DCNN system. DCNN: Deep convolutional neural networks
We also assessed the consistency under the assistance of the DCNN system. We found that the mean inter-observer agreement between the endoscopists was remarkably elevated in seniors (κ: 0.718–0.803 versus 0.773–0.840) and juniors (κ: 0.543–0.664 versus 0.691–0.760) (Fig. 4c, Table S4, Table S5).
Discussion
In this study, we developed a real-time DCNN system to diagnose EGC under NBI. The DCNN system showed a fabulously generalized diagnostic performance with the AUC of 0.947 (95% CI, 0.939–0.956) on the internal validation dataset and the AUCs of 0.888–0.951 on four external validation datasets. The DCNN system also exhibited a robust performance in different subgroups of intraepithelial lesions, intramucosal lesions, and submucosal lesions with the AUCs of 0.915–0.955. On the testing dataset, the diagnostic performance of the DCNN system was superior to that of endoscopists. Notably, the DCNN system markedly elevated the diagnostic performance and inter-observer agreement of senior and junior endoscopists. To facilitate the clinical application of the DCNN system, we also developed an open-access website of the DCNN system for multi-device, including desktop computers, tablets, or smartphones. We also found that the DCNN system could diagnose suspicious lesions in EGD videos in real-time.
Compared with three previous reports of DCNN-based systems, our DCNN system showed several strengths. Firstly, the sample size for the development and validation of our DCNN system was the largest as far as we know. A large amount of data plays a crucial role in developing a robust and generalized DCNN model [20]. Although several strategies have been implemented in developing DCNN models with small datasets, potential risks, including overfitting or low accuracy, may restrain the clinical applicability of the DCNN models [21]. Here, we included a total of 20,208 NBI images to train and validate the DCNN model. With a relatively large dataset, our DCNN model showed a satisfactory diagnostic performance with the AUC, sensitivity, and specificity of 0.947, 98.0%, and 85.2%, respectively. The performance of our DCNN model was quite superior to that of two previous studies with relatively smaller datasets [16, 18]. Intriguingly, another previous report developed a DCNN model with the sensitivity and specificity of 98% and 100%, which achieved a higher specificity [17]. We suppose this may be explained by that our control images were also from the ESD patients. The alteration of background mucosa (including changes in the pits and vascular patterns) may affect the diagnostic accuracy of non-cancer images [22]. Our sensitivity analysis also showed that the abnormality of the background mucosa caused a much lower specificity in submucosal lesions compared with intraepithelial and intramucosal lesions. However, the per-patient analysis showed a marvelous performance of the model, which indicated the relatively low specificity marginally altered the diagnostic accuracy in patient level.
Secondly, we validated the robustness and generalization of our DCNN model comprehensively. We used images from ESD patients with annotations by five experts to train the model with multi-scale predictions in the algorithm. To assess the robustness of our DCNN model, we used temporal validation dataset as the internal validation dataset, performed subgroup analysis based on the invasion depth of lesions, evaluated the real-time performance with EGD videos, and performed human–machine competition. According to the reporting statement, cross-validation and bootstrap validation are often utilized in small-size samples, while temporal validation can be used in large-size samples [23]. Since our dataset is sufficiently large, we split the dataset by time (so-called temporal validation) to ensure the independence of the development and validation dataset. Moreover, our DCNN system diagnosed all the lesions in enrolled 20 consecutive EGD videos, which indicated that our DCNN system might be applicable in the routine real-time EGD examinations. The human–machine competition results showed that our DCNN system's performance was superior to senior and junior endoscopists. Notably, with the assistance of the DCNN system, the diagnostic performance and consistency of endoscopists were remarkably elevated. Notably, we also observed that the accuracy of the combination of the DCNN and endoscopists was lower than that of the DCNN alone. While some studies showed that combined DCNN-human performance could be more strengthened than DCNN alone [24,25,26], other studies also showed that combined DCNN-human performance could not surpass the DCNN performance alone [27,28,29]. We speculated that this may be explained by that some experts could be skeptical of some of DCNN outputs and ignore the recommendations when the predicted results are contradictory to their clinical experience. As a result, it is crucial to build model trust among clinicians to strengthen the collaboration between DCNN and clinicians by presenting the users with easy-to-read manuals, enhancing the explainability of DCNN outputs, and improving the accuracy and generalization of the DCNN outputs [30].
Thirdly, real-time AI systems are often expensive and complicated to be deployed in rural areas. Previous studies have implemented smartphone apps with online AI systems to assist doctors in making diagnosis [31]. Here, we developed an open-access webpage to enable better compatibility for multi-device, by enabling the system to be capable of diagnosing lesions with photographs taken by mobile devices (Fig. 4d, http://112.74.182.39/). This may broaden the application scenarios of the DCNN system to provide convenient and straightforward access for endoscopists in rural and underdeveloped regions.
However, this study has several limitations. Firstly, this was a retrospective study, and the marvelous performance of the DCNN system may not reflect the actual performance in prospective clinical practice. However, we have validated the performance of the DCNN model comprehensively, and this partially demonstrated the robust and stable performance of the DCNN system. Moreover, we have designed a prospective study to validate the performance of our DCNN system. Secondly, we have excluded endoscopic images with poor quality, which may restrain the clinical application of the DCNN system. We are now developing a novel real-time DCNN model to assess image quality and output a score for the image quality, which may facilitate the improvement of endoscopic image quality. Thirdly, the PPVs and NPVs are dependent on prevalence and that the testing image set with 50% prevalence of EGC does not reflect regular clinical practice and therefore the PPV and NPV results may not be the same as real-world performance.
In conclusion, we have developed a real-time DCNN system for EGC diagnosis under NBI and demonstrated that the AI system outperformed the endoscopists and exerted potential assistant impact in EGC identification. However, more prospective validations are needed to evaluate the clinical reinforce of the system in real clinical practice.
Data availability
Due to the privacy of patients, all datasets generated and analyzed in the current study are not available unless a reasonable request to the correspondence author approved by the IRB of Nanjing University Medical School Affiliated Drum Tower Hospital (X.P.Z., zouxp@nju.edu.cn).
References
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics, 2020. CA Cancer J Clin 70:7–30
Katai H, Ishikawa T, Akazawa K, Isobe Y, Miyashiro I, Oda I, Tsujitani S, Ono H, Tanabe S, Fukagawa T, Nunobe S, Kakeji Y, Nashimoto A, Registration committee of the Japanese gastric cancer A (2018) Five-year survival analysis of surgically resected gastric cancer cases in Japan: a retrospective analysis of more than 100,000 patients from the nationwide registry of the Japanese gastric cancer association (2001–2007). Gastric Cancer 21:144–154
Jun JK, Choi KS, Lee HY, Suh M, Park B, Song SH, Jung KW, Lee CW, Choi IJ, Park EC, Lee D (2017) Effectiveness of the Korean national cancer screening program in reducing gastric cancer mortality. Gastroenterology 152(1319–1328):e1317
Hosokawa O, Hattori M, Douden K, Hayashi H, Ohta K, Kaizaki Y (2007) Difference in accuracy between gastroscopy and colonoscopy for detection of cancer. Hepatogastroenterology 54:442–444
Ren W, Yu J, Zhang ZM, Song YK, Li YH, Wang L (2013) Missed diagnosis of early gastric cancer or high-grade intraepithelial neoplasia. World J Gastroenterol 19:2092–2096
Hu YY, Lian QW, Lin ZH, Zhong J, Xue M, Wang LJ (2015) Diagnostic performance of magnifying narrow-band imaging for early gastric cancer: a meta-analysis. World J Gastroenterol 21:7884–7894
Ezoe Y, Muto M, Uedo N, Doyama H, Yao K, Oda I, Kaneko K, Kawahara Y, Yokoi C, Sugiura Y, Ishikawa H, Takeuchi Y, Kaneko Y, Saito Y (2011) Magnifying narrowband imaging is more accurate than conventional white-light imaging in diagnosis of gastric mucosal cancer. Gastroenterology 141(2017–2025):e2013
Florescu DN, Ivan ET, Ciocalteu AM, Gheonea IA, Tudorascu DR, Ciurea T, Gheonea DI (2016) Narrow band imaging endoscopy for detection of precancerous lesions of upper gastrointestinal tract. Rom J Morphol Embryol 57:931–936
Lage J, Pimentel-Nunes P, Figueiredo PC, Libanio D, Ribeiro I, Jacome M, Afonso L, Dinis-Ribeiro M (2016) Light-NBI to identify high-risk phenotypes for gastric adenocarcinoma: do we still need biopsies? Scand J Gastroenterol 51:501–506
White JR, Sami SS, Reddiar D, Mannath J, Ortiz-Fernandez-Sordo J, Beg S, Scott R, Thiagarajan P, Ahmad S, Parra-Blanco A, Kasi M, Telakis E, Sultan AA, Davis J, Figgins A, Kaye P, Robinson K, Atherton JC, Ragunath K (2018) Narrow band imaging and serology in the assessment of premalignant gastric pathology. Scand J Gastroenterol 53:1611–1618
Wu L, Zhang J, Zhou W, An P, Shen L, Liu J, Jiang X, Huang X, Mu G, Wan X, Lv X, Gao J, Cui N, Hu S, Chen Y, Hu X, Li J, Chen D, Gong D, He X, Ding Q, Zhu X, Li S, Wei X, Li X, Wang X, Zhou J, Zhang M, Yu HG (2019) Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut 68:2161–2169
Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M (2020) Deep-learning based detection of gastric precancerous conditions. Gut 69:4–6
Ling T, Wu L, Fu Y, Xu Q, An P, Zhang J, Hu S, Chen Y, He X, Wang J, Chen X, Zhou J, Xu Y, Zou X, Yu H (2020) A deep learning-based system for identifying differentiation status and delineating margins of early gastric cancer in magnifying narrow-band imaging endoscopy. Endoscopy 53(5):469–477
Tang D, Wang L, Ling T, Lv Y, Ni M, Zhan Q, Fu Y, Zhuang D, Guo H, Dou X, Zhang W, Xu G, Zou X (2020) Development and validation of a real-time artificial intelligence-assisted system for detecting early gastric cancer: a multicentre retrospective diagnostic study. EBioMedicine 62:103146
Tang D, Zhou J, Wang L, Ni M, Chen M, Hassan S, Luo R, Chen X, He X, Zhang L, Ding X, Yu H, Xu G, Zou X (2021) A Novel model based on deep convolutional neural network improves diagnostic accuracy of intramucosal gastric cancer (with video). Front Oncol 11:622827
Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C (2020) Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer 23:126–132
Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Matsumoto K, Hojo M, Yao T, Nagahara A, Tada T (2021) Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol 36:482–489
Hu H, Gong L, Dong D, Zhu L, Wang M, He J, Shu L, Cai Y, Cai S, Su W, Zhong Y, Li C, Zhu Y, Fang M, Zhong L, Yang X, Zhou P, Tian J (2020) Identifying early gastric cancer under magnifying narrow-band images with deep learning: a multicenter study. Gastrointest Endosc 93(6):1333-1341.e3
Leisenring W, Alono T, Pepe MS (2000) Comparisons of predictive values of binary medical diagnostic tests for paired designs. Biometrics 56:345–351
Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts H (2019) Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin 69:127–157
Pasini A (2015) Artificial neural networks for small dataset analysis. J Thorac Dis 7:953
Shin N, Jo HJ, Kim W-K, Park W-Y, Lee JH, Shin DH, Choi KU, Kim J-Y, Lee C-H, Sol MY, Jeon TY, Kim DW, Huh GY, Kim GH, Lauwers GY, Park DY (2011) Gastric pit dysplasia in adjacent gastric mucosa in 414 gastric cancers: prevalence and characteristics. Am J Surg Pathol 35(7):1021–1029
Collins GS, Reitsma JB, Altman DG, Moons KG (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350:g7594
Jin EH, Lee D, Bae JH, Kang HY, Kwak MS, Seo JY, Yang JI, Yang SY, Lim SH, Yim JY, Lim JH, Chung GE, Chung SJ, Choi JM, Han YM, Kang SJ, Lee J, Chan Kim H, Kim JS (2020) Improved accuracy in optical diagnosis of colorectal polyps using convolutional neural networks with visual explanations. Gastroenterology 158(2169–2179):e2168
Song Z, Zou S, Zhou W, Huang Y, Shao L, Yuan J, Gou X, Jin W, Wang Z, Chen X, Ding X, Liu J, Yu C, Ku C, Liu C, Sun Z, Xu G, Wang Y, Zhang X, Wang D, Wang S, Xu W, Davis RC, Shi H (2020) Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun 11:4294
Sim Y, Chung MJ, Kotter E, Yune S, Kim M, Do S, Han K, Kim H, Yang S, Lee DJ, Choi BW (2020) Deep convolutional neural network-based software improves radiologist detection of malignant lung nodules on chest radiographs. Radiology 294:199–209
Rajpurkar P, O’Connell C, Schechter A, Asnani N, Li J, Kiani A, Ball RL, Mendelson M, Maartens G, van Hoving DJ, Griesel R, Ng AY, Boyles TH, Lungren MP (2020) CheXaid: deep learning assistance for physician diagnosis of tuberculosis using chest x-rays in patients with HIV. NPJ Digit Med 3:115
Kim HE, Kim HH, Han BK, Kim KH, Han K, Nam H, Lee EH, Kim EK (2020) Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study. Lancet Digit Health 2:e138–e148
Hwang EJ, Lee JS, Lee JH, Lim WH, Kim JH, Choi KS, Choi TW, Kim TH, Goo JM, Park CM (2021) deep learning for detection of pulmonary metastasis on chest radiographs. Radiology 301:455–463
Rajpurkar P, Chen E, Banerjee O, Topol EJ (2022) AI in health and medicine. Nat Med 28:31–38
Zhou W, Yang Y, Yu C, Liu J, Duan X, Weng Z, Chen D, Liang Q, Fang Q, Zhou J, Ju H, Luo Z, Guo W, Ma X, Xie X, Wang R, Zhou L (2021) Ensembled deep learning model outperforms human experts in diagnosing biliary atresia from sonographic gallbladder images. Nat Commun 12:1259
Acknowledgements
We thank Drs. Yanxing Hu, Xianjin Yao (Innovision Medical Technology Co, Xiamen, China), Kaihua Zhang, Yuting Liu, and Zhengwen Wang (Nanjing University of Information Science and Technology) for the development of the DCNN system. Dr. Yanxing Hu described technical methods in the project.
Funding
This project was supported by the National Natural Science Foundation of China (Grant Nos. 81871947), Jiangsu Clinical Medical Center of Digestive System Diseases and Gastrointestinal Cancer (Grant No. YXZXB2016002), and Nanjing Science and Technology Development Foundation (Grant No. 2017sb332019).
Author information
Authors and Affiliations
Contributions
XPZ, GFX, and LW: conceptualized and designed the project. DHT and LW: designed the experiments. MHN, CZ, XWD, NNZ, TY, QZ, YWF, WJL, DMZ, and YL: acquired, curated, and annotated the datasets. DHT: analyzed the data. DHT, GFX, and MHN: wrote the manuscript. XPZ: reviewed and modified the manuscript. All the authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Disclosure
Dehua Tang, Muhan Ni, Chang Zheng, Xiwei Ding, Nina Zhang, Tian Yang, Qiang Zhan, Yiwei Fu, Wenjia Liu, Duanming Zhuang, Ying Lv, Guifang Xu, Lei Wang, and Xiaoping Zou declare no conflict of interest to disclose.
Ethical approval
The present study was approved by the Medical Ethics Committee of Nanjing University Medical School Affiliated Drum Tower Hospital (approval no. 2020–026-01). All procedures were performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (MP4 40257 KB) Real-time detection of the DCNN system for EGC (example 1). DCNN: Deep convolutional neural networks; EGC: Early gastric cancer.
Rights and permissions
About this article
Cite this article
Tang, D., Ni, M., Zheng, C. et al. A deep learning-based model improves diagnosis of early gastric cancer under narrow band imaging endoscopy. Surg Endosc 36, 7800–7810 (2022). https://doi.org/10.1007/s00464-022-09319-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00464-022-09319-2