Abstract
Polyunsaturated fatty acids are important in maintaining human health. Limitations associated with current sources of ω-3 fatty acids and ω-6 fatty acids, from animal and plant sources, have led to increased interest in microbial production. Marine bacteria may provide a suitable alternative, although the isolation of production strains and the identification of operating conditions must be addressed before manufacturing processes become economically viable. Marine isolate 560 was identified as an eicosapentaenoic acid (EPA) producer via GC/MS. The isolate was initially identified as Vibrio cyclitrophicus by 16S rRNA sequencing. Statistically based experimental designs were applied to the optimisation of medium components and environmental factors for the production of EPA. A Plackett–Burman design was used to screen for the effect of temperature, pH, and media components. Subsequently, the concentrations of NaCl, yeast extract, and peptone, identified as significant factors, were optimised using a central composite design. The predicted optimal combination of media components for maximum EPA production (4.8 mg/g dry weight) was determined as 7.9 g/l peptone, 16.2 g/l NaCl, and 6.2 g/l yeast extract. On transfer of this process to bioreactor cultivation, where a range of pH and DO values were tested, the maximum amount of EPA produced increased to 7.5 mg/g dry weight and 10 % of the total fatty acid.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lipids, including fatty acids, are crucial for growth and survival of all organisms. They are important components of membranes and in many organisms they play a significant role in energy storage [1]. ω3 and ω6 polyunsaturated fatty acids (PUFAs) have been associated with cellular and physiological processes where they play an important role in modulating the architecture, dynamics, phase transition, and permeability of membranes and the cohesion of membrane-associated processes [2].
In higher eukaryotes, PUFAs are considered to be associated with fluidity, flexibility, and the selective permeability of the cellular membrane. They also regulate membrane-bound proteins such as ATPase, transport proteins, and histocompatibility complexes [3].
In mammals, the metabolism of PUFA yields an essential group of metabolites known as eicosanoids such as prostaglandins, prostacyclins, thromboxanes, and leukotrienes. This group of metabolites is responsible for regulating many biological processes that help to prevent disorder in mammals by binding to specific G-protein coupled receptors. With such an important role to play in maintaining and regulating whole body functions, PUFAs are referred to as “essential fatty acids” (EFA) [4], needed for normal development and functioning of the mammalian and human body throughout the life cycle.
Biosynthesis of PUFAs by polyketide synthase (PKS)
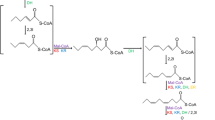
Polyunsaturated fatty acids are generally synthesised by a pathway involving fatty acid synthase (FAS) followed by desaturation and elongation steps from the saturated fatty acid precursors. More recently, Metz et al. [5] described an alternative pathway for the biosynthesis of PUFAs, which uses a polyketide synthase-like (PKS) gene cluster to synthesise PUFAs in both prokaryotes and eukaryotes.
A gene cluster involved in the synthesis of eicosapentaenoic acid (EPA) was first isolated from bacteria by Yazawa [6] from Shewanella sp. strain SCRC-2738 as a genomic fragment of 38 kb cloned into plasmid pEPA in Escherichia coli. Five open reading frames (ORFs), totalling 20 kb, were identified as necessary for the production of EPA in E. coli. Metz et al. [5] identified 11 putative enzymes within the 5 ORFs, 8 of these regions being strongly related to PKS genes and the other three homologues of the bacterial FAS proteins.
Sources of PUFAs
Mammals cannot synthesise these EFA on their own as they lack both of the key enzymes essential for the synthesis of long chain PUFAs de novo and so they have to depend on external dietary sources. Currently, marine fish and plant seed oils are the major sources to fulfil the PUFA requirement in the diet.
The large scale use of marine sources like fish and fish oils for the production of essential fats, which are the potential source of EPA in particular, has been limited due to unacceptable taste and odour, high cholesterol, and small amounts of hazardous toxic impurities [7]. Therefore, the quantity and quality of conventional sources of PUFA may present issues in meeting an increasing market demand. As a result of this, alternative sources are required to overcome these challenges. A variety of PUFAs have been detected in microorganisms including bacteria, fungi, algae, and mosses. Microorganisms are thought to be very promising lipid producers because of their high growth rate on simple media and the simplicity with which they can be manipulated. The diversity of microbial species can facilitate the selection of strains producing lipids with the target fatty acids. Some oleaginous microorganisms have been intensively studied in recent years as alternatives to agricultural and fish oil products [7]. The presence of EPA in the bacterial cells in the form of phospholipids provides a potential added benefit as the application of PUFAs in the form of phospholipids was reported to be more effective than the triglyceride form of PUFAs in various clinical applications for a range of diseases [8, 9].
Past research has revealed a correlation between the distribution of PUFA-producing bacteria and their habitat in deep-sea sediment and cold water [10]. In marine bacteria, it is considered that the low melting temperature of these highly unsaturated membrane components combines with their unique molecular geometry in the membrane, in balancing the competing homeoviscous and homeophasic forces in the cell membrane. PUFA-producing isolates are known to be associated with high pressure, low temperature, deep sea habitats, and are characterised as psychrophilic, halophilic, and predominantly piezophilic [4]. Whilst some research has been carried out in the area of recombinant yeast productions [11], consumer preferences for non-GMO products drive the research in the area of natural producers of PUFA.
PUFA bioprocessing
The aim of this work was to screen marine microorganisms for PUFA production and develop a fermentation process suitable for large scale production. Optimum conditions for growth and production would be identified by initial screening of factors via Plackett–Burman design, followed by a central composite design (CCD) design of experiments. Cultivation in a bioreactor allowed the identification of optimum conditions including physical parameters such as dissolved oxygen (DO) and pH.
Materials and methods
Strains and cultivation conditions
Five different deep sea core sediment and fluff samples were collected from the Mid-Atlantic ridge by research personnel at the Dove Marine laboratory, Newcastle University and kindly provided for this research. After initial screening for PUFA production, isolate 560 was selected for further research. A loopful of cell biomass of this isolate, incubated on bacto marine agar (DIFCO 2216) at 20 °C for 48 h, was transferred to a 250 ml sterile flask containing 50 ml of marine broth and incubated at 20 °C in an orbital shaking incubator at 160 rpm for 2 days. The culture was collected into 50 ml sterile Falcon centrifuge tubes and centrifuged at 4,000g at 4 °C for 15 min. The cell pellets obtained were then transferred into 2 ml sterile centrifuge tubes in 30 % glycerol and stored at −20 °C for subsequent use.
DNA extraction and 16S rRNA amplification
The Gen Elute™ Bacterial Genomic kit from Sigma-Aldrich was used to extract the total genomic DNA of isolate 560. The extracted genomic DNA was used as a template for amplification of the 16S rRNA gene. PCR master mixture, containing 5.0 μl 1× buffer (160 mM (NH4)2SO4, 670 mM Tris–HCl, 0.1 % Tween-20), 0.8 μl a mixture of NTPs, 20 μl each of primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1525r (5′-AAGGAGGTGATCCAGCC-3′) [12], 3.0 μl of 50 mM MgCl2, 0.5 μl of Bio-Taq DNA polymerase (Bioline), and 1 μl of genomic DNA made up with distilled water to 50 μl, was prepared. The PCR products were checked by agarose gel electrophoresis (1 % w/v; 30 min at 100 V, 0.5× TBE). The amplified fragments were compared with 100 bp molecular size marker (MBI Fermentas, Lithuania). The PCR product was stored at −20 °C.
PUFA-PKS-specific gene amplification
Previously extracted genomic DNA was used as a template for the amplification of the specific PUFA PKS gene pfaB using the PCR protocol described in “Strains and cultivation conditions” with the specific degenerative primers pfaB 1240F (GGTGAAGCATCRATGTGGGC) and pfaB 1840R (TCSGCRCCAATTTCAACAA) designed on the basis of the known Shewanella sp. SCRC-2738 (U73935) sequence [13] and the PCR product was checked by agarose gel electrophoresis.
Purification and sequencing of 16S rRNA genes
The PCR product obtained was purified using ExoSAP-IT® mixture (USB, Affymetrix). Following the protocol provided by the supplier, 2 μl of ExoSAP-IT® was mixed with 5 μl of each PCR mixture in a 1.5 ml sterile screw tube, which was then incubated for 15 min at 37 °C, followed by another incubation of 15 min at 80 °C. The purified PCR product was then sent for sequencing (Genius Laboratory, Newcastle upon Tyne, UK).
The quality and quantity of the sequence obtained was checked with Finch TV version 1.4.0. DNA baser (version 3.55.0.199) software was used to assemble the gene. The V4/V5 region [14] was determined in both directions and the full 16S rRNA in draft. The sequence was identified by BLAST and SeqMatch against Genbank and the Ribosomal database project.
Seed culture in artificial sea water
A loopful of biomass from culture plates was transferred into a 250 ml flask containing 50 ml of artificial sea water (ASW) medium (peptone 3.5 g/l; yeast extract 3.5 g/l; NaCl 23 g/l; MgCl2 5.08 g/l; MgSO4 6.16 g/l; Fe2 (SO4)3 0.03 g/l; CaCl2 1.47 g/l; KCl 0.75 g/l; Na2HPO4 0.89 g/l; NH4Cl 5.0 g/l) [15] and allowed to grow at 20 °C in an orbital shaking incubator at 160 rpm for 24 h.
Growth in production media
Growth was performed in 250 ml sterile flasks with 50 ml of given media at given temperature for 2 days in an orbital shaker incubator at 160 rpm. ASW medium was used as the basal media in each experiment. Final biomass from each flask was collected into a 50 ml Falcon centrifuge tube and centrifuged at 4,000g for 15 min. Cell pellets were rinsed with distilled water then re-centrifuged. The cell pellets were transferred into a 1.5 ml screw tube and freeze-dried overnight.
Media optimisation
A Plackett–Burman design of experiments, set up in the Minitab 16 software environment, were used for initial media optimisation experiments. A set of 12 experiments with 8 variables, 7 factors plus a dummy (Table 1) were performed. The initial factors and their minimum and maximum levels were chosen on the basis of the results obtained from preliminary experiments.
The initial Plackett–Burman was followed by a CCD to determine the optimum combination of the most significant factors. A set of 20 experiments with three factors was performed. Final biomass from each flask was collected into a 50 ml Falcon centrifuge tube and processed as described in “Growth in production media”.
The main target of this screening experiment is to compare the significance and the main effect of each factor on the amount of EPA produced by the isolate under investigation. The main effects of each factor were determined by the following equation:
where \( E_{(Xi)} \) is the effect of the tested variable and \( Y_{i}^{ + } \) and \( Y_{i}^{ - } \) are the calculated responses, while the significance level (p value) of each variable was determined using the Student’s t test:
where (SE), the standard error of variables, is calculated as the square root of the variance of an effect. Any variable with (P < 0.1) was considered to be significant at 90 % level of confidence.
The relationship between the response and the significant variables within Plackett–Burman is based on first order polynomial order, as no interactions can be calculated by this screening design.
where Y is the calculated response, \( \beta_{0} \) is model intercept, \( \beta_{i } \) is the regression coefficient for each corresponding variable, \( X_{i } \) is the corresponding variable and k is the number of variables [16].
Effect of ions on growth and EPA production
The metal ions included within the ASW medium were tested via a separate design of experiment. The salts tested were [MgCl2, MgSO4, CaCl2, KCl, Na2HPO4, NH4Cl, and Fe2(SO4)3]. A set of eight experiments with seven variables (Table 2) were performed. The maximum value was the same concentration as ASW medium, while the minimum value was zero.
Optimising CCD
After determining the most significant variables for EPA production, via Plackett–Burman, a CCD was used to estimate the optimum level of each variable. A 20-run design of experiment with each factor at five levels, six replicates at the centre, and star points to estimate the curvature was carried out at 20 °C and pH 8, with basal media containing Na2HPO4, CaCl2, and MgCl2.
A model was parameterised to predict the optimum growth media for EPA production, as a second order polynomial:
where \( \beta_{i } \) are the regression coefficients for each factor, \( \beta_{ii } \) are the regression coefficients for square effect, and \( \beta_{ij } \) are the regression coefficients for interactions. Analysis of variance (ANOVA) was evaluated by statistical analysis of the model by the Design Expert 8.0 statistical package (StatEase, Inc., Minneapolis, MN, USA).
Fatty acid methyl ester (FAME) preparation
Twenty mg of freeze dried cells were suspended in 2 ml of 5 % methanolic HCl and heated at 70 °C for 2 h in sealed tubes. Fatty acid methyl esters were extracted from the cells with 0.6 ml hexane and then dried under nitrogen [17].
FAME profiling
The single point internal standard method was used for the determination of EPA concentration. Methyl nonadecanoate (≥99.5 % GC capillary purity, Sigma-Fluka) was used as an internal standard. Gas chromatography (GC) with flame ionisation detector (FID) on a Hewlett-Packard 5890 series 2 chromatograph, with a SGE forte-BPX70 column; 30 m length and 0.25 μm film thickness from SGE Analytical Science ltd with helium as carrier gas was used for FAME profiling. The GC temperature was held at 210 °C for 30 min (modified and changed from the original method of Jostensen and Landfald [18]).
GC–MS analysis was performed on a Agilent 7890A GC in split mode, injector at 280 °C linked to a Agilent 5975C MSD with the electron voltage 70 eV, source temperature 230 °C, quad temperature 150 °C multiplier voltage 1,800 V, interface temperature 310 °C, controlled by a HP Compaq computer using Chemstation software. The sample (1 μl) in hexane was injected using HP7683B auto sampler with the split open. After the main solvent peak had passed the GC temperature programme and data acquisition commenced. Separation was performed on an Agilent fused silica capillary column (30 m × 0.25 mm) coated with 0.25 μm dimethyl poly-siloxane (HP-5) phase. The GC was temperature programmed from 30 to 130 °C at 5 °C min to 300 °C at 20 °C min and held at the final temperature for 5 min with helium as the carrier gas (flow rate of 1 ml/min, initial pressure of 50 kPa, split at 10 ml/min). Peaks were identified and labelled after comparison of their mass spectra with those of the NIST05 data base.
Bioreactor cultivations
An Ez-Control Applicon bioreactor (working volume 2 l), was used to perform base-line bioreactor cultivations using ASW as culture media. After sterilization, the bioreactor was set up according to the manufacturer’s instructions. 100 ml of the initial seed, grown in ASW medium at 20 °C in an orbital shaking incubator at 160 rpm for 24 h, were added to the vessel. The cultivation temperature was 20 °C, agitation rate was set at 160 rpm and aeration at 2 l of air/min. Samples were taken at 6 h intervals for 3 days and OD600nm measurements were taken. At the end of the cultivation, the bacterial biomass was collected via centrifugation as described in "Growth in production media". FAMEs were extracted and analysed as described in "Effect of ions on growth and EPA production" and "Optimising central composite design". Different combinations of dissolved oxygen (10–30 %) and pH (6–8) were tested.
Results
Strain identification
The 16S rRNA gene sequence analysis initially identified isolate 560 as a Vibrio sp. with a high similarity to the marine type strain Vibrio cyclitrophicus (AM162656), >99 % similarity (1 base difference in the V4/V5 region and 2 bases difference in 1,358 bases of the full 16S sequence (AM162656).
The isolate sequence was submitted to the GenBank database with the accession number (JX203387).
PUFA-PKS-specific gene amplification
To detect the genes associated with PUFA production via the PKS synthesis route, DNA was extracted from the isolate as described in "DNA extraction and 16S rRNA amplification" and used as a template for a PCR reaction using a PUFA-PKS gene primer ("PUFA-PKS specific gene amplification"). Isolate 560 gave a positive result for the presence of pfaB gene.
Identification of the produced EPA
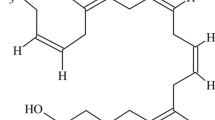
The FAMEs for isolate 560 were analysed by GC/MS to confirm the production of EPA by comparing the mass spectrum with the pure standard. The comparison showed more than 98 % similarity confirming that isolate 560 is an EPA producer (Fig. 1a, b). The amount of EPA produced, in standard ASW medium and prior to optimisation was 1.2 mg/g cell dry weight representing 2 % (w/w) of the total fatty acid.
Media optimisation
Screening Plackett–Burman
Optical density (Y 1 ), EPA concentration (mg/g dry weight) (Y 2 ) and EPA percentage of the total fatty acids (Y 3 ) were used as outputs to be optimised (Table 3). The biomass was harvested after 48 h from inoculation when total EPA was at the maximum level (data not shown).
Statistical analysis of the responses was carried out in Minitab 16 to determine the most significant factors affecting growth and EPA production, 90 % confidence level with α = 0.1 was used in the analysis. A p value equal or less than α was considered significant and the normal plot of the standardised effect revealed if the factor was significant or not (Fig. 2a–c; Table 4).
The first order polynomial models, in terms of the coded factors, were as follows:
-
1.
For optical density,
$$ Y_{1} = { 4}. 5 3 1- 0. 6 10V_{ 1} + 0. 8 5 9V_{ 2} + 0. 8 3 5 { }V_{ 3} - 1. 1 1 1 { }V_{ 4} + \, 0. 5 3 5 { }V_{ 5} + { 1}. 8 2 1 { }V_{ 6} $$ -
2.
For EPA concentration (mg/g dry weight),
$$ Y_{2} = \, 0. 3 { } - 0. 20 6 8 { }V_{ 4} + \, 0. 1 4 6 1V_{ 5} + \, 0. 1 9 6 5 { }V_{ 6} $$ -
3.
For EPA percentage,
$$ Y_{3} = \, 0. 9 { } - 0. 5 3 3 7V_{ 3} - 0. 5 1 5 { }V_{ 4} + \, 0. 5 7 2 3V_{ 5} $$
Higher temperatures were avoided since during preliminary experiments, the amount of EPA dramatically decreased with temperatures above 25 °C and completely disappeared when temperature reached 30 °C (data not shown). To avoid potential cooling problems in large scale production, further experiments were carried out at 20 °C. Yeast extract, peptone and sodium chloride were chosen for the next optimisation step.
Effect of ions on growth and EPA production
The complete absence of the additional metal ions, present in the salts tested, in the growth medium did not affect the growth but led to the complete absence of EPA. The results are summarised in Table 5.
A statistical analysis of the responses was carried out in Minitab 16 to determine the most significant factors affecting growth and EPA production (Fig. 3a–c) and (Table 6).
is as follows:
-
1.
For EPA concentration (mg/g dry weight),
$$ Y_{2} = \, 0. 9 6 8 2+ \, 0. 3 9 9 2C_{ 1} - \, 0. 2 9 6 4C_{ 2} - 0. 3 7 4 1C_{ 3} - 0. 2 4 1 8C_{ 4} + 0. 50 4 4C_{ 5} $$ -
2.
For EPA percentage,
$$ Y_{3} = { 3}.0 9 3 8 { } + \, 0. 6 8 3 7C_{ 1} - \, 0. 7 5 6 3C_{ 2} + \, 0. 4 2 3 7C_{ 3} + 1. 3 5 3 7C_{ 5} - \, 0. 40 1 2C_{ 6} - \, 0. 4 6 1 3C_{ 7}. $$
Central composite design
NaCl, yeast extract and peptone were chosen as factors to establish the optimum medium composition via a CCD design. The matrix and responses of the design are summarised in Table 7.
Analysis of variance of the linear and quadratic effects and the interaction between factors are given in Table 8. Factors with p value <0.1 were considered as significant. Linear and quadratic effects on EPA production seem to be significant, while two way interactions were not.
The 3D surface plots, as a function of two variables, holding the third at the centre level, show the optimal levels of the variables and the relationships between factors (Figs. 4, 5). The optimum combination of factors for maximum production was estimated as NaCl 16.2 g/l, yeast extract 6.6 g/l, and peptone 7.9 g/l. The maximum productivity achieved under these optimum conditions was 4.8 mg of EPA per gram dry weight and represented 6.9 % of the total fatty acids.
Three-dimensional surface plot of EPA concentration (mg/g) as the function of a NaCl and Peptone in un-coded unit (yeast extract was kept at a central point), b peptone and yeast extract in un-coded unit (NaCl was kept at a central point) and c NaCl and yeast extract in un-coded unit (peptone was kept at a central point)
Three-dimensional surface plot of EPA percentage as the function of a NaCl and peptone in un-coded unit (yeast extract was kept at central point), b peptone and yeast extract in un-coded unit (NaCl was kept at central a point) and c NaCl and yeast extract in un-coded unit (peptone was kept at a central point)
Bioreactor cultivation
Batch cultivation in a bioreactor achieved maximum EPA levels after 24–30 h compared to 48 h in shake flasks; this was mainly due to a reduced lag phase.
The bioreactor was run at different combinations of pH and dissolved oxygen (Fig. 6). The optimum EPA concentration and percentage were obtained at high DO and pH 6—approximately 7.5 mg EPA per gram biomass and 10 % of the total fatty acid content.
Discussion
Isolate 560 was initially identified as V. cyclitrophicus through 16S rRNA sequence analysis. Whilst further analyses are required to confirm this identification, the results obtained with this isolate indicate a promising avenue for microbial PUFA production. Optimisation of the media composition for EPA production by the isolate 560 via Placket–Burman and CCD increased the amount of EPA from 1.2 mg/g representing 2 % of the total fatty acids to 4.8 mg/g dry weight representing 7 % of the total fatty acids. By optimising the fermentation conditions, the EPA concentration increased further to 7.5 mg/g dry weight representing 10 % of the total fatty acids.
The Plackett–Burman screening experiment resulted in a number of interesting observations. Glucose appeared to be the most significant factor on growth and EPA productivity but with a negative influence. Although glucose is one of the most widely used carbon sources, it impacted negatively on the growth of the isolate 560. The same effect had been reported on a recombinant E. coli EPA producer as it lost the ability to produce EPA when glucose was added to the media, which could have been due to suppression of the promoter activity [19]. pH also had a significant effect on productivity and growth, with pH 8 resulted in better growth and productivity in shake flask experiment.
In addition to these observations, it was also noted that palmitic acid showed negative effect on EPA production (negative effect value) despite the fact that it is a potential precursor for in vivo production of EPA possibly suggesting the lack of an uptake mechanism. In some microorganisms, the addition of free fatty acids inhibited the biosynthesis of other fatty acids, where the addition of oleic acid to the production media of Phaeodactylum tricornutum showed a remarkable suppression of the biomass and the amount of EPA produced [20]. Higher temperature (20 °C) was preferred for growth (Fig. 2a) while lower temperature (15 °C) was preferred for increased EPA percentage (Fig. 2c) suggesting that lower temperature enhances the ability to produce EPA rather than total fatty acid production. The results of the research reported here indicate that the lower the temperature, the higher the EPA production. This is probably due to the ability of EPA to keep the fluidity of the membrane in cold environment [10]. However, there was no significant effect on EPA concentration in biomass detected when growing isolate 560 at 15 or 20 °C (Fig. 2b).
Yeast extract and peptone played a vital role in the growth and productivity of EPA in this isolate. The complete absence of either of these components during the preliminary experiments resulted in a significant decrease in growth and EPA production. Hedlund and Staley [21] added peptone to the media to initiate the growth of V. cyclitrophicus strain P-2P44 and V. splendidus ATCC 33125 as they observed that an unknown growth factor contained in these is required for growth.
NaCl was chosen for further study due to the marine nature of the isolate to identify the lowest optimum concentration to avoid problems with corrosion due to salt content of media in large scale production.
During the preliminary experiments (data not shown), a wide range of carbon and nitrogen sources (urea, tryptone, ammonium chloride, cellulose, glycerol, glycine, citric acid, sucrose, soy bean flour, ammonium acetate, whey, glutamic acid, molasses, and corn step liquor) did not support the higher production rates achieved with yeast extract or peptone. Although tryptone supported growth, the presence of tryptone in the cultivation media completely inhibited the EPA production. Whilst no reports on similar effect of tryptone on bacteria were found in the literature, Shimiziu et al. [22] reported that, although tryptone yielded a high biomass in the case of Mortierella, the amount of EPA produced was dramatically decreased.
None of the tested ions showed a significant effect on the growth of the isolate under investigation (Fig. 3a). Na2HPO4 showed a significant positive effect on EPA measured as concentration and as percentage of total fatty acids (Fig. 3b, c). This could have been due to the effect of phosphate with EPA present in the bacterial membrane in the form of phospholipid [13]. Na2HPO4 was previously used to test the effect of phosphate on the ability of P. tricornutum to produce EPA [20] showing a significant positive effect on the ability of the isolate under investigation to synthesise EPA.
Magnesium chloride also had a significant positive effect on EPA concentration and percentage (Fig. 3b, c) in line with observations by Akimoto et al. [23]. Bivalent metal ions such as Mg2+ were usually required as cofactors and the enzyme acetyl coenzyme A carboxylase, catalysing the conversion of acetyl coenzyme A to malonyl coenzyme A via a carboxylation step, generally requires such bivalent metal ions [24].
During the Placket–Burman experiments pH 8 was identified as the preferred pH but these flask experiments were performed at relatively low dissolved oxygen, compared to the continuous, high and controlled aeration in the bioreactor. When the DO was low, the growth and the productivity were higher at pH 8 than at pH 6, but increasing the DO in the bioreactor resulted in higher productivity at pH 6 suggesting that there was an interaction between these two physical factors, which affects the EPA production. pH 6 was also reported to be the optimum pH for the EPA producing isolate Shewanella sp. NJ136 [25]. DO appears to play a vital role in EPA production possibly as a result of oxidative pressure, as EPA had been classified as an antioxidant [26, 27]. Since bacterial EPA synthesis was proven to take place via PKAs, which is oxygen-independent pathway [5].
Isolate 560 can be considered to be a potential good producer of EPA, and applying the statistical design of experiment for screening and optimising the production media led to a remarkable enhancement of EPA levels. Following optimisation, isolate 560 was able to produce higher EPA amounts than the Shewanella baltica mutant A, producing 6 mg/g, representing fourfold higher production compared to the wild type [28]. Isolate 560 was found to be able to produce a high percentage of EPA, 10 % of the total fatty acids, at relatively high temperature (20 °C), while other known EPA producing Vibrio species required a low temperature for their maximum EPA production. Vibrio pelagius was able to produce 8.7 % of EPA at 4 °C, but was able to produce only 4.5 % at 23 °C [29]. The amount of EPA produced by the isolate Vibrio sp. NB73 represented 6.4 and 1.3 % of the total fatty acids when the incubation temperature was 10 and 20 °C, respectively [30]. The high percentage of EPA at higher production temperatures observed in isolate 560 is a significant advantage from the large scale manufacture point of view as operating costs can be significantly reduced with lower cooling.
References
Sijtsma L, Swaaf ME (2004) Biotechnological production and applications of the omega-3 polyunsaturated fatty acid docosahexaenoic acid. Appl Microbiol Biotechnol 64(2):146–153
Tapiero H, Nguyen Ba G, Couvreur P, Tew KD (2002) Polyunsaturated fatty acids (PUFA) and eicosanoids in human health and pathologies. Biomed Pharmacother 56(5):215–222
Gill I, Valivety R (1997) Polyunsaturated fatty acids, part 1: occurrence, biological activities and applications. Trends Biotechnol 15(10):401–409
Bergé J, Barnathan G (2005) Fatty Acids from lipids of marine organisms: molecular biodiversity, roles as biomarkers, biologically active compounds, and economical aspects marine biotechnology I. In: Ulber R, Le Gal Y (eds) Advances in biochemical engineering/biotechnology, vol 96. Springer Berlin, Heidelberg, pp 49–125
Metz J, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J (2001) Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293(5528):290–293
Yazawa K (1996) Production of eicosapentaenoic acid from marine bacteria. Lipids 31(1):297–300
Certik M, Shimizu S (1999) Biosynthesis and regulation of microbial polyunsaturated fatty acid production. J Biosci Bioeng 87(1):1–14
Kidd PM (2007) Omega-3 DHA and EPA for cognition, behaviour, and mood: clinical findings and structural–functional synergies with cell membrane phospholipids. Altern med rev 12(3):207–227
Wijendran V, Huang MC, Diau GY, Boehm G, Nathanielsz PW, Brenna JT (2002) Efficacy of dietary arachidonic acid provided as triglyceride or phospholipid as substrates for brain arachidonic acid accretion in Baboon neonates. Pediatr Res 51(3):265–272
Russell NJ, Nichols DS (1999) Polyunsaturated fatty acids in marine bacteria—a dogma rewritten. Microbiology 145(4):767–779
Das T, Thurmond JM, Bobik E, Leonard AE, Parker-Barnes JM, Huang YS, Mukerji P (2000) Polyunsaturated fatty acid-specific elongation enzymes. Biochem Soc Trans 28:658–660
Wawrik B, Kerkhof L, Zylstra GJ, Kukor JJ (2005) Identification of unique type II polyketide synthase genes in soil. Appl Environ Microbiol 71(5):2232–2238
Gentile G, Bonasera V, Amico C, Giuliano L, Yakimov MM (2003) Shewanella sp. GA-22, a psychrophilic hydrocarbonoclastic antarctic bacterium producing polyunsaturated fatty acids. J Appl Microbiol 95(5):1124–1133
Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R (1993) Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res 21(13):3025–3049
Lang S, Hüners M, Lurtz V (2005) Bioprocess engineering data on the cultivation of marine prokaryotes and fungi marine biotechnology II. In: Ulber R, Le Gal Y (eds) Advances in biochemical engineering/biotechnology, vol 97. Springer Berlin, Heidelberg, pp 585–619
Montgomery DC (2000) Design and analysis of experiments, 5th edn. Wiley, Arizona
Watanabe K, Ishikawa C, Yazawa K, Kondo K, Kawaguchi A (1996) Fatty acid and lipid composition of an eicosapentaenoic acid-producing marine bacterium. J Mar Biotechnol 4:104–112
Jostensen JP, Landfald B (1997) High prevalence of polyunsaturated-fatty-acid producing bacteria in arctic invertebrates. FEMS Microbiol Lett 151(1):95–101
Lee SJ, Kim C, Seo PS, Kwon O, Hur BK, Seo JW (2008) Enhancement of heterologous production of eicosapentaenoic acid in Escherichia coli by substitution of promoter sequences within the biosynthesis gene cluster. Biotechnol Lett 30(12):2139–2142
Yongmanitchai W, Ward OP (1991) Growth of and omega-3 fatty acid production by Phaeodactylum tricornutum under different culture conditions. Appl Environ Microbiol 57(2):419–425
Hedlund BP, Staley JT (2001) Vibrio cyclotrophicus sp. nov., a polycyclic aromatic hydrocarbon (PAH)-degrading marine bacterium. Int J Syst Evol Microbiol 51:61–66
Shimiziu S, Kawashima H, Shinmen Y, Akimoto K, Yamada H (1988) Production of eicosapentaenoic acid by Mortierella fungi. J Am Oil Chem Soc 65(9):1455–1459
Akimoto M, Ishii T, Yamagaki K, Ohtaguchi K, Koide K, Yazawa K (1991) Metal salts requisite for the production of eicosapentaenoic acid by a marine bacterium isolated from mackerel intestines. J Am Oil Chem Soc 68(7):504–508
Guchhait RB, Polakis SE, Dimroth P, Stoll E, Moss J, Lane MD (1974) Acetyl coenzyme A carboxylase system of Escherichia coli. J Biol Chem 249(20):6633–6645
Botao Z, Jinlai M, Zhou Z, Guodong W, Quanfu W, Guangyou L, Wanshun L (2007) Screening and optimization of EPA-producing antarctic psychrophilic bacterium Shewanella sp. NJ136. High tech lett 13:95–102
Okuyama H, Orikasa Y, Nishida T (2008) Significance of antioxidative functions of eicosapentaenoic and docosahexaenoic acids in marine microorganisms. Appl Environ Microbiol 74(3):570–574
Nishida T, Morita N, Yano Y, Orikasa Y, Okuyama H (2007) The antioxidative function of eicosapentaenoic acid in a marine bacterium, Shewanella marinintestina IK-1. FEBS Lett 581(22):4212–4216
Amiri-Jami M, Wang H, Kakuda Y, Griffiths M (2006) Enhancement of polyunsaturated fatty acid production by Tn5 transposon in Shewanella baltica. Biotechnol Lett 28(15):1187–1192
Ringo E, Sinclair PD, Birkbeck H, Barbour A (1992) Production of eicosapentaenoic acid (20:5n-3) by Vibrio pelagius isolated from turbot (Scophthalmus maximus (L.)) larvae. Appl Environ Microbiol 58(11):3777–3778
Freese E, Rütters H, Köster J, Rullkötter J, Sass H (2009) Gammaproteobacteria as a possible source of eicosapentaenoic acid in anoxic intertidal sediments. Microb Ecol 57(3):444–454
Acknowledgments
The authors acknowledge the support of the officers and crew of RRS James Cook W. D. K. Reid and Dr. B. Wigham for the provision of sediment samples made available for this study. In addition, authors acknowledge the Egyptian Ministry of Higher Education and Faculty of Science at Mansoura University for funding the research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abd Elrazak, A., Ward, A.C. & Glassey, J. Polyunsaturated fatty acid production by marine bacteria. Bioprocess Biosyst Eng 36, 1641–1652 (2013). https://doi.org/10.1007/s00449-013-0936-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-0936-0