Abstract
Eicosapentaenoic acid (EPA; n-20:5ω3) was found to be a constituent of phospholipids in three mesophilic strains of Gammaproteobacteria, which were isolated from anoxic most probable number series prepared with sediments from an intertidal flat of the German North Sea coast. Their partial 16S rRNA gene sequences identified the isolates as close relatives of Shewanella colwelliana, Vibrio splendidus, and Photobacterium lipolyticum. So far, eicosapentaenoic acid has mainly been reported to occur in eukaryotes and some piezophilic or psychrophilic bacteria. With decreasing temperature, relative contents of EPA (up to 14% of total fatty acids) increased in all strains. Additionally, Shewanella and Vibrio spp. showed a significant increase in monounsaturated fatty acids with lower growth temperature. Analysis of the phospholipid compositions revealed that EPA was present in all three major phospholipid types, namely, phosphatidyl glycerol (PG), cardiolipin and phosphatidyl ethanolamine (PE). However, EPA was enriched in PG and cardiolipin relative to PE. In the tidal flat sediments from which the isolates were obtained, substantial amounts of EPA-containing PG were detected, whereas other typical microeukaryotic phospholipids—being also a possible source of EPA—were abundant at the sediment surface but were present in clearly lower amounts in the anoxic layers beneath 5 cm depth. Therefore, the EPA-containing PG species in the deeper layers in these sediments may indicate the presence of Gammaproteobacteria closely related to the isolates. These bacteria appear to be an important source of EPA in buried, anoxic sediments beneath the layers harboring significant populations of benthic eukaryotes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In bacteria, membranes do not solely act as a cell envelope; they also play a central role in many physiological processes like respiration, chemotaxis, and substrate uptake [23]. These processes rely on a narrow range of membrane phase behavior and organization that may be disturbed by changing environmental conditions [15, 23]. Bacteria are, however, able to alter the composition of their phospholipid fatty-acid side-chains in order to maintain appropriate membrane organization and function [58]. At low temperatures or high pressures, the content of unsaturated fatty acids often increases with a concomitant decline in saturated fatty acids [1, 23]. Polyunsaturated fatty acids (PUFAs), such as eicosapentaenoic acid (EPA; n-20:5ω3) and docosahexaenoic acid (n-22:6ω3), are known to be particularly effective in the adjustment of membrane fluidity due to their low melting points [23, 37]. For a long time, these PUFAs were considered membrane components exclusively of eukaryotes, e.g., unicellular photosynthetic microeukaryotes [60, 61, 66] and fish ([6] and references therein). Accordingly, the presence of PUFAs in sediments was related to eukaryotic organisms [5, 48, 62].
Since the first detection of EPA in marine bacteria by Oliver and Colwell [45] and Johns and Perry [29] in the early to mid-1970s, several EPA-producing bacteria have been isolated. These isolates were generally psychro- or piezophiles from polar regions and the deep sea [10, 13, 20, 40, 44]. However, the recent isolation of mesophilic EPA-producing Shewanella species from a temperate estuary [55] and from shallow seawater samples [19, 27, 28] indicated that EPA may not be restricted to psychrophiles and piezophiles.
All so far known EPA-producing prokaryotes affiliate with only a few genera within two bacterial phyla: The Gammaproteobacteria (e.g., genera Shewanella, Moritella, Colwellia, Alteromonas, and Photobacterium) and the Bacteroidetes (e.g. Flexibacter and Psychroserpens). Therefore, EPA may be a potentially useful marker for these genera in environmental microbial communities if a eukaryotic origin can be excluded. In fact, EPA was detected among the microbial lipids in various polar or deep-sea sedimentary environments (e.g. [9, 12, 39]). However, substantial amounts of polar lipid-bound EPA were also found in temperate regions, e.g., in tidal flat sediments of the German North Sea coast—not only in the surface layers, but down to a depth of 35 cm [51], and more recently down to a depth of a few meters [18]. EPA in these ‘subsurface’ sediment layers was originally assumed to be derived from buried photosynthetic microeukaryotes or protozoan cysts [51].
In the present study, we analyzed three mesophilic bacterial isolates that contain significant amounts of EPA and that were isolated from the aforementioned [51] tidal flat. Bacteria related to these isolates may be a significant source of phospholipid-bound EPA in deeper, anoxic sediments beneath the layers dominated by benthic microeukaryotes. To reveal the potential as source organisms, the intact phospholipids of the isolated bacteria were analyzed and compared to those found in the original sediments. We also analyzed the physiological capacities of the isolates to find out whether they can grow under in situ conditions. To study the role of EPA in adaptation to in situ temperatures, the temperature dependence of fatty- acid patterns of these strains was investigated in detail.
Materials and Methods
Source and Cultivation of Organisms
Pure cultures of marine strains of Gammaproteobacteria were isolated from the highest positive dilutions of most probable number (MPN) dilution series prepared with anoxic media and different carbon substrates (Table 1, Rütters et al., unpublished results). The MPN series were prepared with bicarbonate-buffered and sulfide-reduced artificial seawater as described previously [35, 56]. MPN series were inoculated with sediments from an intertidal flat near the village of Neuharlingersiel (53°42.5′ N, 7°42.05′ E, East Frisian Wadden Sea, NW Germany; [51]) and incubated at 20°C for 6 weeks. Strain SAMA2 was isolated in October 1998 from the upper 0.5 cm of the sediment (T in situ = 12°C) with amino acids (alanine 2.0 mmol·l−1, asparagine 1.0 mmol·l−1, arginine 1.0 mmol·l−1, and cysteine 1.0 mmol·l−1) as substrates. Strains NB72 and NB73 were both obtained from a depth of 5 cm in January 2000 (T in situ = 3°C) with laminarin (0.1%) and chitin (0.1%) as substrates, respectively.
Although the strains were isolated under anoxic conditions, they turned out to be facultative aerobes and, for further investigations, were routinely grown under oxic conditions to achieve higher growth yields. It is expected that this does not affect the production of EPA, as in Bacteria, PUFAs can be synthesized also under anoxic conditions [38, 41] and the fatty-acid patterns between aerobically and anaerobically grown cells are very similar [59]. For fatty-acid analysis, cells were grown in oxic artificial seawater buffered with 4-(2-hydroxyethyl)-1-piperazine-ethanesulfonate [56] using sodium lactate (10 mmol·l−1) as substrate at 10, 20, or 30°C. Close to the extremes of their temperature range for growth, some strains formed only very little biomass, and under the microscope, cells appeared unhealthy. These ‘weak’ cultures were not analyzed for their FA patterns. Since the analysis of intact phospholipids requires a relatively large amount of biomass, several individual cultures (four to eight depending on cell density) were combined before harvest. Parallels were not analyzed because parallel cultures generally show only little variation in their lipid patterns [34]. Cells were harvested at the end of their exponential or during the early stationary growth phase, washed with saline phosphate buffer (9 g l−1 NaCl, 7.5 mmol l−1 NaH2PO4, 7.5 mmol l−1 Na2HPO4, pH 7.2), freeze-dried and stored at −20°C. Intact phospholipids were analyzed from cells grown at 20°C.
Physiological experiments were performed to test the metabolic capacities of the isolates, their ability to grow under anoxic conditions, and the temperature range for growth. Electron donor tests and temperature series were prepared in 22-ml screw-cap tubes filled with 5 ml medium and with air as headspace. Anaerobic electron acceptor utilization and fermentation were tested in completely filled screw cap tubes and analyzed as described previously [35]. For temperature and electron acceptor tests, sodium lactate (10 mmol·l−1) served as electron donor.
16S rDNA Sequencing and Phylogenetic Analysis
The phylogenetic affiliation of the three strains was determined by analysis of partial 16S rRNA genes. Nucleic acids were extracted, and the bacterial 16S rDNA was amplified, using the Bacteria-specific primers 8f and 1492r [36] as described previously [53]. Polymerase chain reaction (PCR) products were purified with the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). Sequence analysis was performed by Amodia Biosystems (Braunschweig, Germany). For phylogenetic analysis, about 550-bp-long fragments were used, as very high similarities (99.3% to 100%) to well described species were obtained, and it was shown that phylogenetic assignments from partial and full-length sequences are very similar [54]. In order to find highest similarities to other bacteria, the novel 16S rDNA sequences of strains SAMA2, NB72, and NB73 were compared using the FASTA3 tool of EMBL-EBI (http://www.ebi.ac.uk). The partial 16S rRNA gene sequences of strains NB72, NB73, and SAMA2 are available at EMBL-EBI under the accession numbers AJ866937 to AJ866939.
Analysis of Whole-Cell Fatty-Acid Patterns
Freeze-dried cells (approximately 20 mg) were saponified under reflux overnight with 5 ml of 5% KOH solution in methanol/water 4:1 v/v under a nitrogen atmosphere. Afterwards, the alkaline solution was decanted into a separation funnel, and the solution was acidified with hydrochloric acid (2 mol·l−1). Non-saponifiable, solid residues were extracted three times with 10 ml dichloromethane each. These dichloromethane extracts were added to the acidified methanol/water phase in the separation funnel. After phase separation, the organic phase was removed, and the aqueous phase was re-extracted three times with 10 ml of dichloromethane each. Combined organic phases were dried over anhydrous sodium sulfate and evaporated to dryness.
Derivatization of Fatty Acids and Analysis by Gas Chromatography
An aliquot of the extracted total fatty acids was transformed into their trimethylsilyl (TMS) esters using N-methyl-N-trimethylsilyltrifluoroacetamide. The TMS derivatives of fatty acids were analyzed on a gas chromatograph (Hewlett Packard HP 5890 series II, Hewlett Packard, Waldbronn, Germany) equipped with a KAS3 cold injection system (Gerstel, Mühlheim a. d. Ruhr, Germany) and a flame ionization detector on a DB-5HT column (30 m × 0.25 mm, 0.1 μm film thickness, J&W, Folsom, CA, USA). For gas chromatography–mass spectrometry (GC-MS), a Finnigan MAT SSQ 710B mass spectrometer (Finnigan-Thermoquest, San Jose, CA, USA) was used. Positions of double bonds were determined using dimethyldisulfide as described by Dunkelblum et al. [16] for selected samples.
Phospholipid and PLFA Analysis
Phospholipid analysis was performed as described in Rütters et al. [52]. In brief, the freeze-dried sediment samples were extracted using a modified Bligh and Dyer procedure [8]. The resulting extracts were separated by liquid chromatography on a silica gel column into various fractions of neutral and polar lipids. The most polar fraction (eluted with dichloromethane/methanol) was analyzed on an high-performance liquid chromatography instrument (Thermo Separation Products, San Jose, CA, USA) coupled to a Finnigan LCQ ion trap mass spectrometer equipped with an electrospray source (Thermoquest-Finnigan, San Jose, CA, USA) and to an evaporative light-scattering detector (Alltech ELSD 500, Deerfield, IL, USA) used for qualitative and quantitative analysis, respectively. Quantification was done after external calibration of ELSD signals with phospholipid standards representing major phospholipid types. Fatty-acid side-chains were assigned according to MS/MS experiments (cf. [51, 52]). Only in the case of cardiolipin the fatty-acid side-chains were only tentatively assigned because not enough information was obtained from MS/MS experiments to rigorously identify all four fatty-acid side-chains. For analysis of polar lipid fatty-acid (PLFA) patterns, an aliquot of the most polar fraction was transesterified by mild alkaline hydrolysis as described by White et al. [64]. The methyl esters obtained were identified by GC-MS (for details see Rütters et al. [52]).
Results and Discussion
Physiology and Phylogeny
The three bacterial strains were isolated under anoxic conditions and grew well by fermentation of amino acids or carbohydrates. Aerobically grown, they utilized a wide range of carbon sources, including biopolymers, carbohydrates, and fatty and amino acids (Table 1). All strains grew with nitrate as electron acceptor. Strain NB72 also reduced manganese oxide and thiosulfate. The temperature range for growth was similar for all strains ranging from 4°C to 35°C, defining them as psychrotolerant mesophiles. With respect to their nutritional capacities and their temperature range for growth, the strains can be seen as well adapted to life in oxic and anoxic sediment layers.
Analysis of partial 16S rRNA genes revealed that the three strains belonged to the Gammaproteobacteria and were closely related to already described species. Strains SAMA2 and NB73 affiliated with the Vibrionaceae, strain SAMA2 being most closely related to Photobacterium lipolyticum T (99.5% sequence similarity, 588 bp) and strain NB73 to Vibrio splendidus T (99.3% sequence similarity, 588 bp). Strain NB72 turned out to be a member of the genus Shewanella within the Altermonadaceae, with a 100% sequence similarity to Shewanella colwelliana T (587 bp).
Fatty-Acid Patterns
Generally, the fatty-acid patterns of the three strains agree well with those of their closest relatives [13, 21, 33, 44, 50, 55], with n-16:0 and n-16:1ω7 being the dominant fatty acids (Table 2). All strains contained relatively high amounts of EPA (up to 14%). The fatty-acid patterns of Photobacterium sp. SAMA2 and Vibrio sp. NB73 were very similar to each other but differed from that of Shewanella sp. NB72 by containing higher amounts of n-16:1ω7 and n-18:1ω7 fatty acids. Shewanella sp. NB72, in turn, possessed much higher amounts of branched-chain fatty acids (10–24%), with i-15:0 being the most prominent one. Trans-fatty acids (mainly trans-16:1ω7) were found in Vibrio sp. NB73, with the highest relative abundance of about 10% in the culture grown at 10°C (Table 2).
Temperature Dependence of Whole-Cell Fatty-Acid Patterns
All three strains changed their fatty-acid patterns with changing temperatures (Table 2, Fig. 1). In Shewanella sp. NB72 and Vibrio sp. NB73, the proportion of monounsaturated fatty acids (MUFAs) increased at lower temperatures (Table 2, Fig. 1), with the most pronounced change occurring between cells grown at 30°C or 20°C. In contrast, in Photobacterium sp. SAMA2, only relatively minor changes in saturated straight-chain fatty acids (SCFAs) and MUFAs were observed, and the ratio of MUFAs to SCFAs remained almost constant (Table 2).
Relative abundances of EPA increased with decreasing growth temperature in all strains. In Shewanella sp. NB72, the relative increase in EPA was highest between cells grown at 30°C to 20°C (max. 5% of total fatty acids). In Vibrio sp. NB73 (maximum, 6.4% of total fatty acids) and Photobacterium sp. SAMA2 (maximum, 13.9% of total fatty acids), largest differences in relative abundances of EPA were observed between cultures grown at 20°C and 10°C. These findings agree with data for other Vibrio [21, 30, 49] and Shewanella species [55, 63]. As in Photobacterium sp. SAMA2, the relative abundances of MUFAs in cells grown at 10°C, 20°C, or 30°C vary only slightly; the relative increase of EPA appears to be the main mechanism of adaptation to low temperatures. Likewise, a mutant strain of Photobacterium profundum produced little MUFAs but elevated levels of EPA and was neither low-temperature nor high-pressure sensitive [1]. Data summarized by Hirota et al. [26] suggested that EPA production of species related to Shewanella pneumatophori is limited to a maximum growth temperature of approximately 30°C, probably indicating that temperature-sensitive enzymes are involved in biosynthesis. This may explain the low values found in Shewanella sp. NB72 grown at 30°C. From the literature, it appears that the amount of EPA produced exclusively depends on temperature but is independent of other environmental parameters like salinity [24], anoxia [38, 41], or variations in nutrient composition and availability, although these were shown to influence the composition of other fatty acids [30].
Branched fatty acids (BCFAs) have repeatedly been reported to promote cold adaptation [4, 11, 58]. However, the results obtained with Shewanella sp. NB72 are ambiguous. Although the relative contribution of BCFAs is higher at 10°C than at 20°C, highest values were observed in cells grown at 30°C. In the latter case, however, it appears that BCFAs compensate for a strong decrease in MUFAs. Similarly, increasing contribution of BCFAs at lower and higher temperatures were observed for Desulfobacterium autotrophicum [46]. For other bacteria, no clear changes in BCFAs with varying growth temperature were observed [25, 43].
Presence of EPA in Bacterial Phospholipids
Major phospholipid types of all three strains were phosphatidyl ethanolamine (PE), phosphatidyl glycerol (PG), and cardiolipin (CL). PE was the most abundant compound in Shewanella sp. NB72 and Vibrio sp. NB73, accounting for 60% and 77% of total phospholipids. These values agree well with literature data [e.g. 14, 19, 45]. In contrast to literature data for other Photobacterium spp. [17, 31, 57], which show a clear dominance of PE, in Photobacterium sp. SAMA2, PE and PG were present in equal amounts accounting for 38% of total phospholipids each.
The most abundant molecular species of all three major phospholipids types possessed combinations of 16:0 and 16:1 fatty-acid side-chains (Table 3). EPA as a side-chain was detected in all three phospholipid types. EPA-containing species were most abundant in PG (17–30% of all PG molecular species) and CL (15–30% of all CL molecular species), in which EPA occurred in combination with saturated and monounsaturated C16 and C18 fatty-acid substituents (Table 3). PE species containing EPA were less abundant (3–10% of all PE molecular species). Furthermore, in PE, EPA was exclusively found in combination with saturated and monounsaturated C16 fatty acids. Even greater differences in the combination of EPA with other fatty acids between PG and PE were reported for Shewanella gelidimarina [42] and a “Vibrio” sp. [24, 25]. Whereas EPA is preferentially combined with BCFAs in PE in these organisms, it is associated with straight-chain saturated or monounsaturated fatty acids in PG.
Presence of EPA in Temperate Tidal Flat Sediments
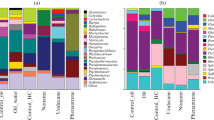
To reveal a potential contribution of the isolates and closely related bacteria to the sediment EPA pool particularly in the anoxic layers, the original sediments were analyzed for their intact phospholipids contents. In the October 1998 core from which Photobacterium sp. SAMA2 was isolated, EPA accounted for up to 15% of the PLFAs. Although absolute amounts of polar-lipid-bound EPA contents generally decreased with depth, even between 10 and 20 cm, i.e., in the anoxic part of the core, up to 4% of the PLFAs were still found to be EPA (data not shown). However, the upper 5 cm and the deeper layers (e.g., 11–15 cm) differed with respect to the EPA-containing intact phospholipid types (Tables 4 and 5): In the uppermost 5 cm of the sediment, EPA was one of the major substituents in phosphatidyl choline (PC), phosphatidyl serine (PS), and phosphatidyl inositol (PI). In addition, PC and PS species with 22:5 and/or 22:6 fatty acids were detected in the uppermost 5 cm. The presence of these typical eukaryotic lipids in the upper anoxic layers (1–5 cm depth) may be explained by the vertical migration of photosynthetic microeukaryotes like diatoms or the presence of other microeukaryotes like ciliates [2, 3, 61, 66]. In contrast, at 11–15 cm depth, phosphatidyl glycerol was found to be the major phospholipid type containing EPA. At this depth, PC was mainly substituted with saturated and monounsaturated C18 fatty acids. C22 PUFAs like those found in microeukaryotes [2, 3, 61, 66] were not detected in any phospholipid type at that depth.
In the January 2000 core, from which Shewanella sp. NB72 and Vibrio sp. NB73 were isolated, 8% and 3% of the PLFAs were found to be EPA at the surface and at a depth of 2–6 cm, respectively. As in the October 1998 core, EPA was one of the major fatty-acid side-chains in PC and PI at the sediment surface, with smaller portions being present in PG and PE. However, already at a sediment depth of 2–6 cm, the detected phospholipid species indicate that the proportion of eukaryotic biomass is low. The most abundant molecular species among PC contained two 18:1 fatty-acid substituents, while relative abundances of EPA-containing phospholipid species remained nearly constant among PG (Tables 4 and 5).
Gammaproteobacteria as a Source of EPA in Anoxic Sediment Layers
Since PUFA-producing bacteria were so far known only from polar regions or deep-sea habitats, it was assumed that buried algal biomass was the source of EPA detected in ‘subsurface’ sediment layers (cf. [5, 7]). However, in the phospholipids of photosynthetic microeukaryotes, PUFAs with 16, 18, 20, and 22 carbon atoms occur in addition to EPA (e.g., [2, 3, 32]). In our sediments, PUFAs other than EPA were only detected among the phospholipids in the uppermost centimeters of sediment, but not in deeper layers (Table 5). Furthermore, in deeper sediment layers, the major EPA-containing phospholipid type was phosphatidyl glycerol, a common bacterial phospholipid, whereas contents of PC and PI that are major microeukaryotic lipids [2, 3, 32] decreased rapidly with depth in both sediment cores (Table 4). Therefore, photosynthetic microeukaryotes such as diatoms are the most likely source of EPA-containing phospholipids in the surface sediments, whereas EPA bound to PG detected in deeper sediment layers indicates a bacterial origin. Gammaproteobacteria related to the genera Photobacterium, Vibrio, and Shewanella such as our isolates appear to be promising candidates as source organisms because PG is one of their major EPA-containing phospholipid types. However, contents of PE, the most abundant phospholipid type in our isolates, were surprisingly low in deeper sediments. This may be explained by the contribution of other EPA-producing bacterial genera possessing PG as their major phospholipid type. Molecular surveys of sediments from the area have revealed a large diversity of so far uncultured Bacteria including Gammaproteobacteria only remotely related to any known genera [65]. However, the assumption that bacteria closely related to the three isolates investigated in this study are abundant in the tidal flat sediments, and hence, a likely source of EPA is supported by the repeated isolation from highest dilutions of MPN series ([35], Sass and Rütters, unpublished results] but also by molecular investigations. Hardwick et al. [22] showed that members of the genera Vibrio (up to 28%), Shewanella (up to 6%), and related Gammaproteobacteria can represent a significant fraction of the microbial communities in coastal sediments.
Using the conversion factors presented by Rajendran et al. [47], which are 100 μmol PLFAs per·gram bacterial dry weight and 5.9 × 1012 cells per·gram dry weight, MPN counts given in Table 1 can be converted into PLFA contents. The estimated potential contribution of EPA by our isolates ranges from 0.0006 (Photobacterium sp. SAMA2, October 1998) to 0.078 μg EPA g−1 sediment (Vibrio sp. NB73, January 2000). These values are close to those found in situ. In the October 1998, core EPA contents in the PLFA fraction were 0.54 μg EPA g−1 sediment in the 1- to 5-cm-depth interval [51] in which a significant contribution of eukaryotes was found as indicated by the high contents of PC and PI (Table 4). In contrast, in the 11- to 15-cm-depth interval, which was dominated by bacterial lipids, only 0.09 μg EPA g−1 sediment was found [51]. This indicates that bacteria may indeed be the main source of EPA in the layers underneath the zone harboring photosynthetic microeukaryotes. Since all three strains were isolated under anoxic conditions and grew well by fermentation and were capable of using alternative electron acceptors, it can be expected that they are actively growing and present in numbers also at greater depths.
Conclusions
The presence of eicosapentaenoic acid in anoxic sediment layers has previously been attributed to buried algal cells, microeukaryotes, or burrowing zoobenthos. The presence of EPA in bacterial lipids was rather seen as a physiological curiosity restricted to psychrophiles from polar regions or the deep sea. However, the occurrence of EPA in our investigated strains of Shewanella, Vibrio, and Photobacterium spp. suggests that the ability to produce PUFAs is more widespread among mesophilic Gammaproteobacteria than previously known. As these strains were isolated from anoxic layers and grew well anaerobically, our results suggest that Gammaproteobacteria are the likely source of EPA present in deeper, anoxic sediments, particularly, as it has been shown that various bacteria can synthesize PUFAs under strictly anoxic conditions (e.g., [38, 41]). Analysis of the intact phospholipids shows that, at the sediment surface, EPA is present in lipids typically found in photosynthetic microeukaryotes, whereas in the anoxic layers, EPA is mostly present in PG, as in our bacterial isolates. These results indicate that the interpretation of the presence of algal cells in sediment based on the occurrence of EPA has to be done with caution. Analysis of intact phospholipids can provide supporting information to allow a better distinction between eukaryotic and bacterial microbial community members.
References
Allen EE, Facciotti D, Bartlett DH (1999) Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl Environ Microbiol 65:1710–1720
Alonso DL, Belarbi EL, Rodriguez-Ruiz J, Segura CI, Gimenez A (1998) Acyl lipids of three microalgae. Phytochemistry 47:1473–1481
Anderson R, Livermore BP, Kates M, Volcani BE (1978) The lipid composition of the non-photosynthetic diatom Nitzschia alba. Biochim Biophys Acta 528:77–88
Annous BA, Becker LA, Bayles DO, Labeda DP, Wilkinson BJ (1997) Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 63:3887–3894
Belicka LL, MacDonald RW, Yunker MB, Harvey RH (2004) The role of depositional regime on carbon transport and preservation in Arctic Ocean sediments. Mar Chem 86:65–88
Bell MV, Henderson RJ, Sargent JR (1986) The role of polyunsaturated fatty acids in fish. Comp Biochem Phys B 83:711–719
Blair NE, Levin LA, DeMaster DJ, Plaia G (1996) The short-term fate of fresh algal carbon in continental slope sediments. Limnol Oceanogr 41:1208–1219
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Bowman JP, McCammon SA, Gibson JAE, Robertson L, Nichols PD (2003) Prokaryotic metabolic activity and community structure in Antarctic continental shelf sediments. Appl Environ Microbiol 69:2448–2462
Bowman JP, McCammon SA, Nichols DS, Skerratt JH, Rea SM, Nichols PD, McMeekin TA (1997) Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5ω3) and grow anaerobically by dissimilatory Fe(III) reduction. Int J Syst Bacteriol 47:1040–1047
Chattopadhyay MK, Jagannadham MV (2001) Maintenance of membrane fluidity in Antarctic bacteria. Polar Biol 24:386–388
DeLong EF, Yayanos AA (1985) Adaptation of the membrane lipids of a deep-sea bacterium to change in hydrostatic pressure. Science 228:1101–1103
DeLong EF, Yayanos AA (1986) Biochemical function and ecological significance of novel bacterial lipids in deep-sea prokaryotes. Appl Environ Microbiol 51:730–737
De Siervo AJ, Reynolds JW (1975) Phospholipid composition and cardiolipin synthesis in fermentative and nonfermentative marine bacteria. J Bacteriol 123:294–301
Dowhan W (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66:199–232
Dunkelblum E, Tan SH, Silk PJ (1985) Double bond location in monounsaturated fatty acids by dimethyl disulfide derivatisation and mass spectrometry: application to analysis of fatty acids in pheromone glands of four Lepidoptera. J Chem Ecol 11:265–277
Eberhard A, Rouser G (1971) Quantitative analysis of the phospholipids of some marine bioluminescent bacteria. Lipids 6:410–415
Freese E, Köster J, Rullkötter J (2008) Origin and composition of organic matter in tidal flat sediments from the German Wadden Sea. Org Geochem 39:820–829
Frolova GM, Pavel KG, Shparteeva AA, Nedashkovskaya OI, Gorshkova NM, Ivanova EP, Mikhailov VV (2005) Lipid composition of novel Shewanella species isolated from far Eastern Seas. Microbiology 74:664–669
Gentile G, Bonasera V, Amico C, Giuliano L, Yakimov MM (2003) Shewanella sp. GA-22, a psychrophilic hydrocarbonoclastic Antarctic bacterium producing polyunsaturated fatty acids. J Appl Microbiol 95:1124–1133
Hamamoto T, Takata N, Kudo T, Horikoshi K (1995) Characteristic presence of polyunsaturated fatty acids in marine psychrophilic Vibrios. FEMS Microbiol Lett 129:51–56
Hardwick EO, Ye W, Moran MA, Hodson RE (2003) Temporal dynamics of three culturable γ-Proteobacteria taxa in salt marsh sediments. Aquat Ecol 37:55–64
Hazel JR (1995) Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu Rev Physiol 57:19–42
Henderson RJ, Millar R-M, Sargent JR (1995) Effect of growth temperature on the positional distribution of eicosapentaenoic acid and trans hexadecenoic acid in the phospholipids of a Vibrio species of bacterium. Lipids 30:181–185
Henderson RJ, Millar R-M, Sargent JR, Jostensen J-P (1993) Trans-Monoenoic and polyunsaturated fatty acids in phospholipids of a Vibrio species of bacterium in relation to growth conditions. Lipids 28:389–396
Hirota K, Nodasaka Y, Orikasa Y, Okuyama H, Yumoto I (2005) Shewanella pneumatophori sp. nov., an eicosapentaenoic acid-producing marine bacterium isolated from the intestines of Pacific mackerel (Pneumatophorus japonicus). Int J Syst Evol Microbiol 55:2355–2359
Ivanova EP, Sawabe T, Gorshkova NM, Svetashev VI, Mikhailolov VV, Nicolau DV, Christen R (2001) Shewanella japonica sp. nov. Int J Syst Evol Microbiol 51:1027–1033
Ivanova EP, Sawabe T, Hayashi K, Gorshkova NM, Zhukova NV, Nedashkovskaya OI, Mikhailov VV, Nicolau DV, Christen R (2003) Shewanella fidelis sp. nov., isolated from sediments and sea water. Int J Syst Evol Microbiol 53:577–582
Johns RB, Perry GJ (1977) Lipids of the marine bacterium Flexibacter polymorphus. Arch Microbiol 114:267–271
Jostensen J-P, Landfald B (1996) Influence of growth conditions on fatty acid composition of a polyunsaturated-fatty-acid-producing Vibrio species. Arch Microbiol 165:306–310
Kalacheva GS, Vysotskii ES, Rodicheva EK, Fish AM (1981) Lipids of the luminescent bacteria Photobacterium mandapamensis. Microbiology 50:56–60
Kates M, Volcani BE (1966) Lipid components of diatoms. Biochim Biophys Acta 116:264–278
Kato C, Nogi Y (2001) Correlation between phylogenetic structure and function: examples from deep-sea Shewanella. FEMS Microbiol Ecol 35:223–230
Könneke M, Widdel F (2003) Effect of growth temperature on cellular fatty acids in sulphate-reducing bacteria. Environ Microbiol 5:1064–1070
Köpke B, Wilms R, Engelen B, Cypionka H, Sass H (2005) Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl Environ Microbiol 71:7819–7830
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 113–175
Marsh D (1990) Handbook of lipid bilayers. CRC, Boca Raton
Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domerque F, Yamada A, Yazawa K, Knauf V, Browse J (2001) Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293
Nichols DS (2003) Prokaryotes and the input of polyunsaturated fatty acids to the marine food web. FEMS Microbiol Lett 219:1–7
Nichols DS, Bowman J, Sanderson K, Nichols CM, Lewis T, McMeekin T, Nichols PD (1999) Developments with Antarctic microorganisms: culture collections, bioactivity screening, taxonomy, PUFA production and cold-adapted enzymes. Curr Opin Biotechnol 10:240–246
Nichols DS, Nichols PD, McMeekin T (1992) Anaerobic production of polyunsaturated fatty acids by Shewanella putrefaciens strain ACAM 342. FEMS Microbiol Lett 98:117–122
Nichols DS, Nichols PD, Russell NJ, Davies NW, McMeekin TA (1997) Polyunsaturated fatty acids in the psychrophilic bacterium Shewanella gelidimarina ACAM 456T: molecular species analysis of major phospholipids and biosynthesis of eicosapentaenoic acid. Biochim Biophys Acta 1347:164–176
Nichols DS, Presser KA, Olley J, Ross T, McMeekin TA (2002) Variation of branched-chain fatty acids marks the normal physiological range for growth in Listeria monocytogenes. Appl Environ Microbiol 68:2809–2813
Nogi Y, Masui N, Kato C (1998) Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2:1–7
Oliver JD, Colwell RR (1973) Extractable lipids of gram-negative marine bacteria: fatty acid composition. Int J Syst Bacteriol 23:442–458
Rabus R, Brüchert V, Amann J, Könneke M (2002) Physiological response to temperature changes of the marine, sulfate-reducing bacterium Desulfobacterium autotrophicum. FEMS Microbiol Ecol 42:409–417
Rajendran N, Matsuda O, Urushigawa Y (1992) Distribution of polar lipid fatty acid biomarkers for bacteria in sediments of a polluted bay. Microbios 72:143–152
Rajendran N, Nagatomo Y (1999) Seasonal changes in sedimentary microbial communities of two eutrophic bays as estimated by biomarkers. Hydrobiologia 393:117–125
Ringø E, Sinclair PD, Birkbeck H, Barbour A (1992) Production of eicosapentaenoic acid (20:5 n-3) by Vibrio pelagius isolated from Turbot (Scophthalmus maximus (L.)) larvae. Appl Environ Microbiol 58:3777–3778
Russell NJ, Nichols DS (1999) Polyunsaturated fatty acids in marine bacteria—a dogma rewritten. Microbiology 145:767–779
Rütters H, Sass H, Cypionka H, Rullkötter J (2002) Microbial communities in a Wadden Sea sediment core—clues from analyses of intact glyceride lipids, and released fatty acids. Org Geochem 33:803–816
Rütters H, Sass H, Cypionka H, Rullkötter J (2002) Phospholipid analysis as a tool to study microbial communities. J Microbiol Meth 48:149–160
Sass A, Rütters H, Cypionka H, Sass H (2002) Desulfobulbus mediterraneus sp. nov., a sulfate-reducing bacterium growing on mono- and disaccharides. Arch Microbiol 177:468–474
Schmidt TM, DeLong EF, Pace NR (1991) Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol 173:4371–4378
Skerratt J, Nichols PD, Bowman JP (2002) Shewanella olleyana sp. nov., a marine species isolated from a temperate estuary which produces high levels of polyunsaturated fatty acids. Int J Syst Evol Microbiol 52:2101–2106
Süß J, Engelen B, Cypionka H, Sass H (2004) Quantitative analysis of bacterial communities from Mediterranean sapropels based on cultivation-dependent methods. FEMS Microbiol Ecol 51:109–121
Süß J, Herrmann K, Seidel M, Cypionka H, Engelen B, Sass H (2008) Two distinct Photobacterium populations thrive in ancient Mediterranean sapropels. Microb Ecol 55:371–383
Suutari M, Laakso S (1994) Microbial fatty-acids and thermal adaptation. Crit Rev Microbiol 20:285–328
Teece MA, Fogel ML, Dollhopf ME, Nealson KH (1999) Isotopic fractionation associated with biosynthesis of fatty acids by a marine bacterium under oxic and anoxic conditions. Org Geochem 30:1571–1579
Volkman JK, Brown MR, Dunstan GA, Jeffrey SW (1993) The biochemical composition of marine microalgae from the class Eustigmatophyceae. J Phycol 29:69–78
Volkman JK, Dunstan GA, Jeffrey SW, Kearney PS (1991) Fatty acids from microalgae of the genus Pavlova. Phytochemistry 30:1855–1859
Volkman JK, Johns RB (1977) The geochemical significance of positional isomers of unsaturated acids from an intertidal zone sediment. Nature 267:693–694
Wang F, Wang P, Chen M, Xiao X (2004) Isolation of extremophiles with the detection and retrieval of Shewanella strains in deep-sea sediments from the west Pacific. Extremophiles 8:165–168
White DC, Davis WM, Nickels JS, King JD, Bobbie RJ (1979) Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oecologia 40:51–62
Wilms R, Sass H, Köpke B, Köster J, Cypionka H, Engelen B (2006) Specific eubacterial, archaeal and eukaryotic communities in tidal flat sediments along a vertical profile of several meters. Appl Environ Microbiol 72:2756–2764
Zhukova NV, Aizdaicher NA (1995) Fatty acid composition of 15 species of marine microalgae. Phytochemistry 39:351–356
Acknowledgments
The authors acknowledge the technical assistance by Dipl.-Ing. B. Kopke and would like to thank John K. Volkman for helpful discussion in the forefront of this paper. We thank six anonymous referees for their constructive comments and support. This work is a part of the research group on “BioGeoChemistry of Tidal Flats” and was funded by Deutsche Forschungsgemeinschaft (DFG grant no. RU 458/33).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Freese, E., Rütters, H., Köster, J. et al. Gammaproteobacteria as a Possible Source of Eicosapentaenoic Acid in Anoxic Intertidal Sediments. Microb Ecol 57, 444–454 (2009). https://doi.org/10.1007/s00248-008-9443-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9443-2