Abstract
The marine strain Pseudomonas otitidis was isolated to hydrolyze the cooked sunflower oil (CSO) followed by the production of lipase. The optimum culture conditions for the maximum lipase production were determined using Plackett–Burman design and response surface methodology. The maximum lipase production, 1,980 U/ml was achieved at the optimum culture conditions. After purification, an 8.4-fold purity of lipase with specific activity of 5,647 U/mg protein and molecular mass of 39 kDa was obtained. The purified lipase was stable at pH 5.0–9.0 and temperature 30–80 °C. Ca2+ and Triton X-100 showed stimulatory effect on the lipase activity. The purified lipase was highly stable in the non-polar solvents. The functional groups of the lipase were determined by Fourier transform-infrared (FT-IR) spectroscopy. The purified lipase showed higher hydrolytic activity towards CSO over the other cooked oil wastes. About 92.3 % of the CSO hydrolysis was observed by the lipase at the optimum time 3 h, pH 7.5 and temperature 35 °C. The hydrolysis of CSO obeyed pseudo first order rate kinetic model. The thermodynamic properties of the lipase hydrolysis were studied using the classical Van’t Hoff equation. The hydrolysis of CSO was confirmed by FT-IR studies.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lipids (characterized as oils, greases and fats) are one of the most important components of natural foods and many synthetic compounds and emulsions. Therefore, the lipids are widely found in municipal and industrial waste waters. Used edible oils (cooked oil wastes) are also considered as a problematic waste, contributing to the environmental pollution. The treatment of lipid-rich wastewater is still a challenge. Even though lipids are biogenic compounds, their different structures and solubilities imply that it may be difficult to induce the enzymes required for their biodegradation. Therefore, the use of viable microorganisms is necessary because they have to subsist in the lipid substrate and induce the enzyme (lipase) to hydrolyze them to glycerol and fatty acid, and further biodegrade them to carbon dioxide and water.
Marine microorganisms could able to provide an interesting alternative for the degradation of lipids, due to their unique genetic structures and life habitats when compared to the terrestrial microorganisms [1]. The marine environment ranges from nutrient-rich regions to nutritionally sparse locations where only a few organisms can survive. With the recent advent of biotechnology, there has been a growing interest and demand for enzymes with novel properties. The complexity of the marine environment involving high salinity, high pressure, low temperature and special lighting conditions, may contribute to the significant differences between the enzymes generated by marine microorganisms and homologous enzymes from terrestrial microorganisms, leading to the boosted marine microbial enzyme technology in recent years and the resulting valuable products. Because of the novel properties of enzymes from marine microorganisms, they can able to catalyze various biochemical reactions (including hydrolysis) [2]. However, there are very few reports available on their production, characterization and application of the enzymes from marine microorganisms [3].
Apart from the biodegradation of lipids using the marine lipolytic microorganisms, the recovery of high value-added product (lipase) from the lipid-fermented medium is considered to be the viable method for using the lipase for the hydrolysis of lipids in large scale operations. Lipase is one of the most extensively used enzymes, it can able to hydrolyse the triglycerides into glycerol and fatty acids at the lipid–water interface systems [4]. Lipases are widely used in several areas of medicinal, biotechnological, detergent, dairy, food, leather, textile, surfactant, pharmaceutical and hydrolysis of fats and oil processing owing to their substrate specificity such as fatty acid, alcohol and region-stereospecificity [5, 6]. The most important application of lipase in oleochemical industry is the production of fatty acids from oils through hydrolysis [7].
An efficient strategy for enhancing yield of the desired metabolic product in a microbial system is the optimization of medium components [8]. In recent years, use of statistical approach involving Plackett–Burman (PB) designing and response surface methodology (RSM) has gained lot of impetus for medium optimization and for understanding the interactions among various physico-chemical parameters using a minimum number of experiments [9–11]. Response surface methodology uses the quantitative data from appropriate experiments to determine and simultaneously solve multivariate equations [12, 13].
In the present study, the cooked sunflower oil was used as the model substrate for the biodegradation of municipal waste water containing lipids and industrial lipid wastes using lipase. To degrade/hydrolyse the CSO, and to produce the novel lipase, highly efficient marine lipolytic strain P. otitidis was isolated from the marine sediment. Amongst the various bacterial lipases that are being exploited, those from the genera Pseudomonas have unique properties such as temperature stability, high enantio-selectivity and activity in broader range of pH [14]. It is for this reason that an extensive research has been done on lipase production, using Pseudomonas sp. Therefore, the strain P. otitidis was used for the fermentation of lipid substrate followed by the production of lipase. The culture conditions were optimized using PB designing and RSM. The produced lipase was purified and characterized. The purified lipase was applied as a biocatalyst for the hydrolysis of different cooked waste oils such as cooked sunflower oil (CSO), cooked olive oil (COO), cooked palm oil (CPO), cooked coconut oil (CCO), cooked groundnut oil (CGO), and cooked gingelly oil (CGiO). The hydrolytic rate kinetic constants and thermodynamic properties were evaluated for the hydrolysis of CSO using the lipase from P. otitidis using CSO.
Materials and methods
Materials
The cooked sunflower oil prepared from the sunflower oil was obtained from a local restaurant, Chennai, India. The composition of the CSO was linoleic acid (66 %), oleic acid (32 %), palmitic acid (11 %), and stearic acid (9 %).
Bacterial strain isolation and cultivation
Lipase producing microbial cultures were isolated from the sediments of about 10–20 m from seashore (Kovalam beach (77′15″N, 8′05″E), Chennai). Therefore, 1 g of sediment was serially diluted in the sterile seawater and the marine isolates were isolated using artificial seawater agar (Himedia) using the pour plate method. The pure culture was screened by their lipolytic activity on Tributyrin agar plates, grown at 37 °C. Based on their zone of hydrolysis, the lipolytic microorganism was selected and maintained on double-strength nutrient agar slants and glycerol and stored at 4 °C.
Lipolytic bacterial strain identification
The strain, which showed maximum lipolytic activity, was identified by 16S ribosomal DNA (16S rDNA) sequencing and phylogenetic analysis. The genomic DNA was isolated according to the procedure of Marmur [15]. To identify the strain UNI16SF (5′-GAG AGT TTG ATC CTG GC-3′) and UNI16SR (5′-AGG AGG TGA TCC AGC CG-3′) primers were used for amplification of the variable region of the 16S rDNA gene. Phylogenetic analysis was performed by subjecting the deduced sequence to the 16S rDNA database to obtain the similar sequences and the phylogenetic tree was constructed using the Phylip package [16].
Lipase production
Optimization experiments
Lipase was produced by growing the culture in a marine medium containing sunflower oil 10 ml/L. The same medium with 1 % sunflower oil supplemented culture was used to optimize lipase production. Lipase production was optimized by varying the time (0–120 h), pH (5.5–8), temperature (20–60 °C), external carbon sources (dextrose, sucrose, maltose, lactose and fructose), and concentration of substrate (1, 2, 3, 4, 5 and 10 %). The various significant factors were screened using PB design and then optimized using response surface design.
Plackett–Burman design
The preliminary experiments revealed that five factors, including time, temperature, concentration of substrate, pH and external carbon source, were supposed to have effect on lipase production. It was known that the PB design could evaluate the main effects of factors. The factors having significant effects on lipase production were identified using this experimental design. As shown in the Table 1, the design matrix covered five factors with two dummy variables to evaluate their effects on lipase production and lipase activity was given as response value. Each factor was investigated at a high (+1) and a low (−1) level. The factors, which were significant at 95 % of confidence level (P < 0.05) were considered to have greater effects on the lipase production and further optimized using response surface methodology.
The first order model used to fit the results of PB design was represented as Eq. (1)
where Y was the predicted response, β 0 was the intercept, β i was the linear coefficient and X i was the coded independent factor.
Response surface methodology
Response surface methodology was used to optimize the most significant factors for improving lipase production, screened by PB design. The four independent factors were studied at three different levels and a set of 30 experiments were carried out.
The model equation used for the analysis was given as Eq. (2)
where Y is the predicted response, k is the number of factors, α 0 is the design factor of interest, α i and α ij are coefficients. The accuracy and general ability of the above polynomial model could be evaluated by the coefficient of determination R 2. Each experimental design was carried out in duplicate, and the mean values were given.
Statistical analysis
Design expert, version 8.0.7 (Statease Inc., Minneapolis, USA) was used for the experimental designs and regression analysis of the experimental data. Statistical analysis of the model was performed to evaluate the analysis of variance (ANOVA). The quality of the polynomial model equation was judged statistically by the coefficient of determination R 2 and its statistical significance was determined by an F test.
Experimental validation of the optimized condition
In order to validate the optimization of medium composition, three tests were carried out using the optimized conditions to confirm the result from the analysis of the response surface.
Optimum culture conditions
Pseudomonas otitidis isolated from marine sediment was grown in Zobell marine broth (Himedia) with 1 % CSO as the substrate. The substrate was emulsified in broth, using an ultrasonicator (BANDELIN, Germany) at 23 kHz. Culture conditions were 300 ml of medium in one litre Erlenmeyer flasks, rotation at 100 rpm at 37 °C. Biomass in the broth was removed by centrifugation and the supernatant was subjected to lipase activity determination.
Enzyme activity assay
Lipase activity was measured by titrimetric assay, using an olive oil emulsion [17] with slight modifications. Olive oil 10 % (v/v) was emulsified in distilled water containing 2 % (w/v) of polyvinyl alcohol in an ultrasonicator (BANDELIN, Germany) at 23 kHz. Lipase assay was determined by adding 5 ml of olive oil emulsion, 2 ml of 0.03 % Triton X-100, 2 ml of 3 M NaCl, 1 ml of 0.075 % CaCl2 and 5 ml of phosphate buffer to the 1 ml of enzyme. The enzyme–substrate mixture was incubated at 37 °C for 15 min. The reaction was terminated by adding ethanol: acetone (1:1, v/v) to the mixture. The liberated fatty acids were titrated against 0.02 N NaOH, using phenolphthalein as indicator. The sample containing the reaction mixture and ethanol:acetone solution without the addition of enzyme was titrated against 0.02 N NaOH and was used as control. One activity unit of lipase was defined as the amount of enzyme that released 1 μmol of fatty acid per minute under assay conditions.
Purification of lipase
The enzyme from the cell free supernatant was precipitated by ammonium sulphate at 80 % saturation at 4 °C. The precipitate was collected by centrifugation at 8,000g for 15 min at 4 °C and dissolved in 0.1 M phosphate buffer of pH 7.5. This aliquot was dialyzed against the same buffer overnight under refrigerated conditions. The quantitative activity of the lipase was determined as previously described. The dialyzed enzyme was loaded onto the DEAE-cellulose column (1.0 × 15 cm) previously equilibrated with the 0.1 M phosphate buffer (pH 7.5). After washing in the two bed volumes of the initial buffer, elution was performed with a negative linear gradient of 0–1.0 M NaCl at a flow rate of 30 ml/h. Fractions showing lipase activity were collected and lyophilized for further analysis.
Determination of protein and molecular weight
Protein concentration was determined by Lowry method [18] employing bovine serum albumin as the standard. The molecular mass of the lipase was determined by using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) according to the method followed by Laemmli [19], on a 5 % stacking gel and 12 % resolving gel. The protein marker ranging from 14.3 to 94.7 KDa was used as a standard marker for determination of molecular weight. Protein bands were visualized by silver staining.
Determination of optimum reaction conditions
Effect of pH and temperature on activity and stability of purified lipases
The optimum pH for the purified lipase was determined by incubating the enzyme substrate at various pH ranging from 5 to 9 at room temperature in the following buffers: 0.1 M acetate buffer (5.0, 5.5, 6.0 and 6.5), 0.1 M phosphate buffer (7.0, 7.5 and 8.0) and 0.1 M Tris buffer (8.5 and 9). The residual enzyme activity was determined, using the standard assay as described previously. The pH stability was studied by incubating the lipase at each desired pH for 4 h and then the lipase activity was measured.
The optimum temperature for the purified lipase was determined by incubating the reaction mixture at temperatures ranging from 20 to 80 °C at optimized pH value. The thermal stability was studied by incubating lipase mixture at each temperature for 4 h and lipase activity under same assay conditions was measured.
Effect of metal ions, detergents, inhibitors and organic solvents
One millimole of KCl, CaCl2, ZnCl2, MgCl2, FeSO4, CuSO4, MnCl2, and EDTA was added to 0.1 M phosphate buffered (pH 7.5) enzyme solution at 35 °C for 1 h to determine the stimulatory or inhibitory effects of metal ions. Similarly, different detergents such as 0.1 % (v/v) of Triton X-100, Tween 20, Tween 80 and 1 mM of SDS and inhibitor such as β-mercaptoethanol were incubated with the buffered enzyme (pH 7.5) at 35 °C for 1 h. The effect of various organic solvents on the enzyme was carried out by the addition of 10 and 20 % of polar solvents such as acetone, ethanol, methanol, isopropanol, dimethyl formamide (DMFA), dimethyl sulphoxide (DMSO), and acetonitrile and non-polar solvents such as benzene, hexane, pentane, octane, and toluene for 1 h at 35 °C. After the incubation period, the lipase activity was measured using the titrimetric assay.
Determination of kinetic parameters
The olive oil and cooked sunflower oil emulsions at different concentrations (1–10 %), at a constant temperature (35 °C) and pH (7.5) were employed to determine the kinetic parameters such as maximum reaction rate (V max) and Michaelis–Menton constant (K m) for the purified lipase. One unit of lipase activity (U) is defined as the amount of enzyme that hydrolyzed the substrates to release 1 μmol of fatty acid per minute. The kinetic parameters were obtained from Lineweaver–Burk equation plot Eq. 3
where [S] is the substrate concentration (mM) and V is the initial reaction rate of the enzyme (mM/min).
Fourier transform-infrared spectroscopy (FT-IR) of purified lipase
The functional groups present in the purified lipase were identified using a Perkin Elmer FT-IR spectrophotometer. For this purpose the samples were made in the form of pellet with the thickness of 1 mm and diameter of 13 mm, using spectroscopic grade KBr. The spectrum was analyzed in the spectral range of 400–4,000 cm−1.
Kinetic studies on the hydrolysis of CSO using the purified lipase from P. otitidis using CSO
The kinetic studies were evaluated to determine the efficiency of lipase produced from CSO on the hydrolysis of CSO. The optimum conditions for the hydrolysis of CSO were carried out by varying the time (15–180 min), pH (3.0–9.0) and temperature (20–50 °C) in the batch experiment. The experiment was carried out in a 100 ml conical flask containing the 15 ml of substrate (CSO) emulsion which was prepared as follows: one ml of CSO was added with the 15 ml of 0.1 M phosphate buffer at pH 7.5 and it was emulsified using ultrasonicator (BANDELIN, Germany) at 23 kHz. The enzyme (5,647 U or 1 mg) was added with the substrate emulsion. The enzyme substrate mixture was incubated for the hydrolysis reaction at 35 °C. The samples were withdrawn and analyzed the residual lipid content according to the method followed by Joseph et al. [20]. The % conversion of CSO was calculated as follows.
Nonlinear kinetic model for the hydrolysis of CSO using the purified lipase.
In order to investigate the kinetic rate constants for the hydrolysis of CSO using the purified lipase from P. otitidis, the non-linear kinetic models were performed. The pseudo first order [21] and pseudo second order [22] were employed with following equations (Eqs. 5 and 6), respectively.
where r e and r t are the amount of CSO hydrolyzed (mg/mg of lipase) at equilibrium and at time (t), K 1 and K 2 are the first and second order rate constants.
Thermodynamic studies on the hydrolysis of CSO using purified lipase from P. otitidis using CSO
The classical Van’t Hoff equation was used to determine the thermodynamic properties such as Gibbs free energy change, entropy and enthalpy of hydrolysis of CSO using purified lipase. A change in temperature would mean that only the Eq. (7) could be used.
Hydrolysis of different cooked waste oils by purified lipase from P. otitidis using CSO.
The cooked waste oils (CWO) such as cooked olive oil (COO), cooked palm oil (CPO), cooked coconut oil (CCO), cooked groundnut oil (CGO), cooked sunflower oil (CSO), and cooked gingelly oil (CGiO) were obtained from different restaurants and used as a substrate for the purified lipase. One ml of each CWOs were added with the 15 ml of 0.1 M phosphate buffer at pH 7.5 and it was emulsified using ultrasonicator (BANDELIN, Germany) at 23 kHz. The enzyme (5,647 U or 1 mg) was added with the substrate emulsion. The enzyme substrate mixture was incubated for 3 h at 35 °C. After the incubation period, the samples were withdrawn and analyzed the residual lipid content [20]. The % conversion of CWO was calculated using the Eq. (4).
FT-IR studies for the functional group determination of hydrolyzed products of CSO
The functional groups of the unhydrolyzed and hydrolyzed CSO using purified lipase from P. otitidis using CSO as the substrate were identified using a Perkin Elmer FT-IR spectrophotometer. For this purpose the samples were lyophilized and made in the form of pellet with the thickness of 1 mm and diameter of 13 mm, using spectroscopic grade KBr. The spectrum was analyzed in the spectral range of 400–4,000 cm−1.
Results and discussion
Isolation and identification of lipolytic microorganism
Among 25 strains isolated from the marine sediments, only one strain showed the maximum lipolytic activity (144 U/ml) and a clear zone formation on tributyrin agar plates indicating the production of an extracellular lipase and thus, the strain was selected for the production of lipase. The 16S rDNA sequencing data indicated that the isolate was P. otitidis (Fig. 1). The nucleotide sequence reported here has been assigned from NCBI Gene bank database. The assigned accession number is JQ638704 The BLAST result showed 98 % similarities to other P. otitidis strain MCC10330 (accession number: NR 043289.1). Pseudomonas otitidis is an aerobic, gram negative bacterium which grows at pH 7.5 and temperature 35 °C.
Plackett–Burman design
Plackett–Burman design offered an effective screening procedure and evaluated the significance of a large number of factors in one experiment, which was time-saving. Table 1 shows the coded values of high and low level of factors chosen for the trials in PB design. Table 2 represents the five independent factors and two dummy variables with their concentrations at different coded levels and the experimental responses for eight runs.
Statistical analysis of the data showed that time, temperature, pH and external carbon source had a significant effect on lipase activity (Table 2). The F test confirms the significance of factors and the factors having values greater than 0.5 could be considered as significant.
Response surface methodology (RSM)
Following screening, RSM using central composite design was employed to determine the optimal levels of the four selected factors that significantly affected lipase activity. The high and low levels with the coded levels for the factors are as same as in PB design. Based on the regression analysis of the data from the Table 3, the effects of four factors on lipase activity were predicted by the second order polynomial function as
where A, B, C, D are time, pH, temperature and dextrose, respectively.
Analysis of variance (ANOVA) for response surface quadratic model
ANOVA (partial sum of squares—type III)
The statistical significance of equation was checked by F test and ANOVA for the second order polynomial model as shown in Table 3. The analysis of factor (F test) showed that, the second order polynomial model was well adjusted to the experimental data and the coefficient of variation (CV), indicated the degree of precision of the experiment. In general, higher the value of CV, lower the reliability of the experiment. Here a lower value of CV (10.14) indicated a better precision and reliability of experiments [23]. The precision of a model can be checked by the regression coefficient (R 2). The regression coefficient was calculated to be 0.9726, indicating that the 97.26 % of the variability in the response could be explained by this model. Linear and quadratic terms were both significant at the 1 % level. Therefore, the quadratic model was selected in this optimization study.
The regression coefficients and corresponding P values are given in Table 3. The P values were used as the tool to check the significance of each coefficient, which was necessary to understand the pattern of the mutual interactions between the best factors. Smaller the P value, bigger the significance of the corresponding coefficients [24–26]. The results showed that, among independent factors, A (time) and B (pH) had more significant effects on lipase activity.
Values of “Prob > F” less than 0.0500 indicate model terms are significant. In this case A, B, AB, AC, AD, A2, B2, C2, D2 are significant model terms. Values greater than 0.1000 indicate the model terms are not significant.
Comparison of observed and predicted lipase activity
A regression model could be used to predict the future observations on the response (lipase activity) corresponding to particular values of the regressor variables. In predicting new observations and in estimating the mean response at a given point, one must be careful about extrapolating beyond the region containing the original observations. It was very possible that a model that fitted well in the region of the original data would no longer fit well outside the region. In this experiment, the predicted data of the response from the empirical model Eq. (8) was in good agreement with the observed ones in the range of the operating variables.
Localization of the optimum condition
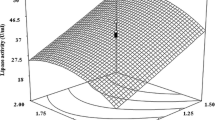
The 3D response surface plots described by the regression model were drawn to illustrate the effects of the independent factors and interactive effects of each independent factor on the response factors. And also it showed the optimum conditions required for the maximum production of lipase was time 75 h, temperature 35 °C, pH 7.5, and dextrose concentration 1.25 g/L (Fig. 2). Each figure presented the effect of two factors while the other factor was held at the zero level.
At the optimized condition (time 75 h, temperature 35 °C, pH 7.5, and dextrose concentration 1.25 g/L), and at the 4 % substrate concentration, the maximum lipase activity was found to be 1,980 U/ml. The production of lipase depends on the optimum culture conditions of the respective microorganisms. The same explanation was given by Ghosh et al. [27]. They reported that the best temperature for the lipase production and the growth in the case of R. nigricans was 30 °C while for T. emersonii, it was around 45 °C.
Purification of lipase
The culture supernatant was used as a starting material for the purification of lipase. The lipase was purified by ammonium sulphate precipitation and DEAE-cellulose column chromatography. The lipase was recovered as a single peak in void volume. Table 4 summarizes the results of lipase purification. About 8.4-fold purification with 19.4 % recovery was achieved. Gaur et al. [28] reported that the purification fold and recovery of other Pseudomonas sp. lipases were in the range of 13.9–34.7-fold and 2.9–16.4 %, respectively. The specific activity of the purified lipase was found to be 5,647 U/mg protein. The molecular mass of the protein was found to be 39 kDa based on SDS-PAGE (Fig. 3). The position of single band in non-denaturing PAGE (Fig. 3; lane 2) indicates possible formation of aggregates [29]. Most of the known Pseudomonas sp. lipases have been reported to have molecular mass in the range of 29–33 kDa with a few reports on higher molecular mass in the range of 45–94 kDa [28, 30–32].
Characterization of purified lipase
Effect of pH and temperature on lipase activity and stability
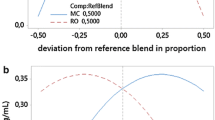
The purified lipase from P. otitidis was able to tolerate a broad range of pH 6.0–9.5 (Fig. 4a). The maximum relative lipase activity was observed at pH 7.5. However, beyond pH 9.5 (at pH 10), the relative lipase activity was reduced to 22 %. The purified lipase was incubated for 1 h in different pH to determine the stability of the purified lipase over the acidic, neutral and alkaline pHs. The study showed the lipase possessed good stability over wide range of pHs. About 72–83 % of the relative lipase activity was stable in the acidic pH range 5.0–6.0 and 90 % of the relative lipase activity was observed at neutral pH (pH 7.5). About 85–55 % of the relative lipase activity was maintained after 1 h incubation in the pH range of 8–9.5. The data on pH optimum and pH stability proves the P. otitidis lipase was found to be alkaline in nature. Almost the similar trend was observed for the other Pseudomonas sp. lipases reported by researchers [33, 34].
The purified lipase was active in the studied range of temperature from 20 to 60 °C with a higher activity at 35 °C (Fig. 4b). The decline in relative lipase activity was observed at the temperatures of 70 and 80 °C, respectively with the relative lipase activity of 45 and 30 %. This could be attributed due to the loss of catalytic activity taking place at these temperatures. The lipases derived from other Pseudomonas sp. such as P. fluorescens 2D, P. fluorescens HU380, P. fragi and P. mendoncina were also found to be optimally active within 35 to 45 °C [33, 34]. While studying the thermal stability profile, the purified lipase was highly stable (100 %) at 35 °C. The purified enzyme maintained 58 and 78 % of its relative lipase activity at 20 and 80 °C, respectively. The results showed that the lipase was stable in the broad range of temperature. Due to this potential property, the lipase could be applied in wide industrial applications.
Effect of metal ions and reducing agent
Nearly one-third of all known enzymes require the presence of metal ions for catalytic activity [35]. This group of enzymes includes the metal enzymes, which contain tightly bound metal ion cofactors, most commonly transition metal ions such as Fe2+, Cu2+, Mn2+, and Zn2+. Metal-activated enzymes, in contrast, loosely bind metal ion from solution, usually the alkali and alkaline earth metal ions K+, Mg2+, and Ca2+. Among the metal ions tested, lipase activity was strongly enhanced by Ca2+ ion (relative lipase activity: 112 %) (Table 5). The other metal ions such as Mg2+ and K+ showed slight inhibiting effect on the relative lipase activity about 83 and 45 %, respectively. The rest of the metal ions were acted as a strong inhibitor for the lipase activity. The stimulatory effect on the lipase activity by Ca2+ suggests that the enzyme needed calcium ion as a catalytic activator. Moreover, the results suggest that the lipase activity was enhanced by the addition of alkali and alkaline earth metal ions than the transition metal ions. Hence, the P. otitidis lipase is found to be a metal-activated enzyme. In this group of enzymes, the ions often play a structural role rather than a catalytic one. The ions bind to the enzyme and change the conformation of the protein to counter greater stability to the enzyme. But transition metal ions change the confirmation of the protein to less stable due to ion toxicity. The addition of chelating agent, EDTA showed inhibitory effect (there was only 27 % of the relative lipase activity is remaining) suggesting that the lipase purified from P. otitidis is a metallo enzyme.
Effect of detergents and reducing agent on lipase activity
The stability of lipase in surfactant is desirable for lipases to remain active in detergent formulations [36]. Surfactants facilitate the access of substrate to the lipase by stabilizing the interfacial area where catalytic reaction of lipase takes place [37]. The addition of non-ionic/anionic surfactant can increase both the activity and the stability of surface active enzymes. The effect of non-ionic and anionic surfactants on lipase activity was studied for 1 h duration at 35 °C and pH 7.5 by choosing the surfactants with hydrophilic–lipophilic balance (HLB) varying in an interval of 13–16.7. The HLB value indicates that the polarity of the molecule in arbitrary units with its value increases with the increase in hydrophilicity [38]. The addition of Triton X-100 increased the relative lipase activity (118 %) (Table 5), which had a HLB value of 13.0. It was interesting to note that the relative lipase activity decreased to 52 and 32 % for Tween 80 (HLB-15.0) and Tween 20 (HLB-16.7), respectively, when increasing the HLB values. The decrease in relative lipase activity with increasing HLB values of Tween series may be due to the fact that the excessive adsorption of the surfactants on the enzyme surface results in diffusional limitation on the reaction. On contrary, the anionic surfactant sodium dodecyl sulphate (SDS) owing to its negative charge showed an inhibitory effect towards the relative lipase activity (12 %) as it possessed a HLB value 40.
Results in Table 5 also show the role of disulphide linkages in lipase catalysis. The disulphide bond has an important role in stabilizing the protein structure. The Pseudomonas lipase has two cysteine residues, which form a disulphide bond [39]. Exposure of P. otitidis lipase to S–S reducing agent β-mercaptoethanol reduced the relative enzyme activity to 92 % indicating importance of disulphide bridges [37].
Stability towards organic solvents
Employing lipases for the bioconversions in organic solvents is advantageous from biotechnological point of view. Hence, stability in solvents is considered as novel attribute in a lipase. Generally, enzymes, being proteins, lose their activity at organic solvent concentrations higher than 10–20 % [40]. Therefore, the stability of lipase on various organic solvents was tested at 10 and 20 % solvent concentrations (Table 5).
The lipase was stable in the presence of polar solvents (log P < −1.378) like DMSO, methanol, ethanol and acetone as well as non-polar hydrophobic solvents ranging from log P (2.13–4.783). The non-polar solvent octane showed better stimulatory effect (248 % of relative lipase activity) than the other non-polar and polar solvents. In the overall study, the lipase is very stable in non-polar solvents than the polar solvents. It is reported that polar solvents strip off the essential water molecules from the active site of enzymes. For this reason, use of polar solvents is avoided and hydrophobic solvents are more often employed in non-aqueous enzymology. The non-polar solvent tolerant lipases therefore appear promising for catalysis. The stability of the lipase in aqueous–organic mixtures suggested the ability of this enzyme to resist denaturation by organic solvents and to form multiple hydrogen bonds with water for structural flexibility and conformational mobility for optimal catalysis [41]. Therefore, the marine P. otitidis lipase is naturally stable in organic solvent and holds the potential for use in organic synthesis and related applications [42].
Enzyme kinetic parameters
Kinetic parameters, Km and vmax for P. otitidis lipase were determined by Lineweaver–Burk plot (Fig. 5). The Km and vmax values of purified lipase were carried out using olive oil and CSO substrates. The results showed that the Km and vmax values for olive oil substrate were found to be 20.715 and 1.034 mM/min, respectively. The Km and vmax values for CSO substrate were found to be 7.324 and 2.309 mM/min, respectively. The Km value represents higher affinity between enzymes and substrates while vmax represents the higher catalytic efficiency of lipase [43]. Our results were corroborated with the other Pseudomonas lipases [31, 44].
Functional groups of purified lipase analyzed by FT-IR
The FT-IR spectra in Fig. 6 show major protein bands owing to the peptide group vibration that occurred in the spectral region 1,200–1,700 cm−1. The band at 1,652.38 cm−1 is due to the C=O stretching vibrations of amide I. The band at 1,233.33 and 1,461.52 cm−1 can be attributed to NH bending and C–N stretching vibrations [45]. The other Pseudomonas lipase reported also showed the band at 1,652 cm−1 is due to the C=O stretching vibrations of amide I. The band at 1,238 and 1,406 cm−1 can be attributed to NH bending and C–N stretching vibrations [30].
Kinetic studies on the hydrolysis of CSO using the purified lipase
Many different bacterial species produce lipases to hydrolyze esters of glycerol, preferably with long-chain fatty acids. These lipases act at the interface generated by a hydrophobic lipid substrate in a hydrophilic aqueous medium. In the present study, the hydrolysis of CSO was studied using the lipase derived from P. otitidis using CSO as the substrate. The reaction is as follows. 
Effect of time, pH and temperature on the hydrolysis of CSO using purified lipase
The lipase was reacted with the CSO substrate in the presence of phosphate buffer (pH 7.0). The batch hydrolysis of CSO with respect to time is shown in Fig. 7a. The maximum percentage conversion of CSO (58.4 %) was achieved at 3.0 h, thereafter the hydrolysis reaction attained saturation. Thus, the optimum time was considered to be 3.0 h in the present investigation.
Effect of a time (conditions: concentration of CSO 33.3 ml/L, pH 7.0 and temperature 30 °C). b pH (conditions: concentration of CSO 33.3 ml/L, time 3 h and temperature 30 °C) and c temperature (conditions: concentration of CSO 33.3 ml/L, time 3 h and pH 7.5) on the hydrolysis of CSO by lipase produced from CSO
The pH on the hydrolysis of CSO using lipase at 30 °C was carried out for 3 h as shown in Fig. 7b. Various pH buffer solutions from 3.0 to 9.0 were prepared, using acetate, phosphate and tris buffers. The results revealed that the maximum conversion of CSO (65.4 %) was obtained at pH 7.5. The percentage conversion of CWO was decreased when the pH value was below or above 7.5. This was correlated with the optimum pH for the purified lipase activity, which was observed a pH 7.5. At this pH, the lipase was highly active and thus the maximum hydrolysis of CSO also attained at the same pH (7.5).
The hydrolysis reaction rates of lipase at different temperatures (20–50 °C) are shown in Fig. 7c. The optimum temperature for the hydrolysis of CSO using lipase was 35 °C with 92.3 % of the hydrolysis.
Nonlinear kinetic model for the hydrolysis of CSO using the purified lipase
The first order rate constant k 1 and the second order rate constant k 2 are summarized in Table 6. Moreover, the results confirmed that the hydrolysis of CSO by the lipase purified from P. otitidis obeyed the first order rate kinetic model as observed greater R 2 values.
Thermodynamic studies on the hydrolysis of CSO using purified lipase from P. otitidis using CSO
The thermodynamic parameters were calculated using the Eq. (7). The negative change in entropy value (ΔS = −45.208 J/molK) indicates that the hydrolysis of CSO using purified lipase has become more ordered. Moreover, the positive value of enthalpy (ΔH = 14.913 kJ/mol) of the system indicates the hydrolysis of CSO using purified lipase is an endothermic process. The positive free energy (ΔG = 28.611 kJ/mol) value indicate the process of hydrolysis of CSO using purified lipase is not spontaneous.
Hydrolysis of different cooked waste oils by purified lipase from P. otitidis using CSO
It has been reported that the hydrolytic activity is the basic characteristic of lipase. However, the same lipase has different hydrolytic activity towards different kinds of lipids from different sources. It is required that lipase is non-specific with high specific activity, hydrolyzing different kinds of lipids from different sources when it is applied to lipase biosensor, detergent industry and digestion of lipids in the food and medicine [46]. In order to determine the specificity of the lipase from P. otitidis using CSO, the lipase was used for the hydrolysis of different lipid substrates such as CSO, palm oil, gingelly oil, groundnut oil, and coconut oil. Interestingly, the study showed the higher hydrolytic efficiency (92 %) towards CSO (Fig. 8). This may be explained due to the substrate specificity of the lipase. Apart from CSO, the lipase showed better hydrolytic efficiency towards cooked palm oil (85 %), groundnut oil (82 %), coconut oil (73 %), and gingelly oil (68 %). The results suggested that lipase used in the present study had good hydrolytic efficiency towards non-specific substrate with more specificity towards their own substrate. Therefore, the lipase with high hydrolytic activity towards different CWOs may have high potential applications in degradation of lipids from different industries.
Identification of hydrolyzed products by FT-IR
The FT-IR spectrum of unhydrolyzed CSO (Fig. 9a) shows O–H stretching vibrations at 3,368.55 cm−1. The C=O stretching of ester peak was observed at 1,739.25 cm−1. The –CH2 scissoring vibrations was observed at 1,456.06 cm−1 and the C–O stretching vibration was found in the region 1,088.21 cm−1.
The FT-IR spectrum of hydrolyzed CSO (Fig. 9b) by lipases shows that there was a shifting of C=O stretching of esters peak from lower region to higher, i.e., from 1,739.25 to 1,746.26 cm−1, which is due to the cleavage of triglycerides in the CSO into glycerol and fatty acids. Moreover, the spectrum clearly confirms the formation of glycerol and fatty acids from the triglyceride form of CSO. This could be explained in detail as follows. The band observed at 1,361.90 and 1,152.38 cm−1 is due to the –C–O stretching vibration of carboxylic acid overlapped with –OH bending vibrations of glycerol and –CH2 wagging, respectively.
Conclusions
The main objective of the study was to produce the high yield of lipase from marine bacterial strain P. otitidis by utilizing CSO as the substrate. The strain was identified by using 16S rDNA sequencing and phylogenetic analysis. The strain P. otitidis produced 1,980 U of lipase activity per milliliter of the fermented medium at the optimum conditions, which was comparatively higher than the other lipases produced from all other microorganisms. The optimum culture conditions were determined by using PB design and response surface methodology. The purified lipase was active in broad pH range 5.0–9.0 and temperature range 30–80 °C. Ca2+ ions and non-ionic surfactant Triton X-100 showed stimulatory effect on the purified lipase activity. The non-polar solvent octane showed better stimulatory effect than the other non-polar and polar solvents studied. The amide groups of purified lipase were identified by FT-IR spectroscopy. The purified lipase showed better hydrolytic activity towards CSO than the other substrates used. This shows the substrate specificity of the lipase which was produced using CSO as the substrate. About 92.3 % of the CSO hydrolysis was observed at the optimum conditions. The hydrolysis of CSO followed the pseudo first order rate kinetic model. The thermodynamic study indicated the process of hydrolysis of CSO using purified lipase is not spontaneous. The FT-IR studies confirmed the fragmentation of triglyceride form of CSO into glycerol and fatty acid. The attractive characteristics of lipase from marine strain P. otitidis using CSOs and their higher hydrolytic efficiency towards the lipid substrates will be very useful for the wide variety of industrial applications including the hydrolysis of oils and fats.
References
Stach JEM, Maldonado LA, Ward AC, Goodfellow M, Bull AT (2003) New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ Microbiol 5:828–841
Ranjitha P, Karthy ES, Mohankumar A (2009) Purification and characterization of the lipase from marine Vibrio fischeri. Int J Biol 1:48–56
Bhatnagar T, Boutaiba S, Hacene H, Cayol JL, Fardeau ML, Ollivier B, Baratti JC (2005) Lipolytic activity from Halobacteria: screening and hydrolase production. FEMS Microbiol Lett 248:133–140
Ramani K, John Kennedy L, Ramakrishnan M, Sekaran G (2010) Purification, characterization and application of acidic lipase from Pseudomonas gessardii using beef tallow as a substrate for fats and oil hydrolysis. Process Biochem 45:1683–1691
Gombert AK, Pinto AL, Castilho LR, Freire DMG (1999) Lipase production by Penicillium restrictum in solid-state fermentation using babassu oil cake as substrate. Process Biochem 35:85–89
Maia MMD, Heasley A, Camargo de Morais MM, Melo EHM, Morais MAJ, Ledingham WM (2001) Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresour Technol 76:23–27
Shamel MM, Ramachandran KB, Hasan M, Al-Zuhair S (2007) Hydrolysis of palm and olive oils by immobilized lipase using hollow-fibre reactor. Biochem Eng J 34:228–235
Pankaj S, Manpreet S, Ashwini LK, Uttam CB (2007) Response surface optimization of the critical medium components for carbonyl reductase production by candida viswanthii MTCC 5158. Bioresour Technol 98:829–833
Gupta N, Mehra G, Gupta R (2004) A glycerol inducible thermostable lipase from Bacillus sp.: medium optimization by a Plackett–Burman design and by response surface methodology. Can J Microbiol 50:361–368
Kalil SJ, Maugeri F, Rodrigues MI (2000) Response surface analysis and simulation as a tool for bioprocess design and optimization. Process Biochem 35:539–550
Rathi P, Goswami VK, Sahai V, Gupta R (2002) Statistical medium optimization and production of a hyper thermostable lipase from Burkholderia cepacia. J Appl Microbiol 93:930–936
Gorret N, Rosli SK, Oppenheim SF (2004) Bioreactor culture of oil palm (Elaeis guineensis) and effects of nitrogen source, inoculum size and conditioned medium on biomass production. J Biotechnol 108:253–263
Tokcear Z, Bayraktar E, Mehmetoglu U (2006) Response surface optimization of antidipteran delta-endotoxin production by Bacillus thuringiensis subsp. israelensis HD 500. Process Biochem 41:350–355
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 64:763–781
Marmur JA (1961) Procedure for the isolation of deoxyribonucleic acids from microorganisms. J Mol Biol 3:208–218
Felsenstein J, Phylip (1993) Phylogenetic inference package, version 3.51c. Bacteriology 98:756–766
Naci D, Ali AM (2002) Partial purification of intestinal triglyceride lipase from Cyprinion macrostomus heckel, 1843 and effect of pH on enzyme activity. Turk J Biol 26:133–143
Lowry OH, Rosebrough NJ, Farr AL, Randal J (1951) Protein measurement with the Folin-phenol reagent. J Biol Chem 193:265–275
Laemmli UK (1970) Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680–685
Joseph AK, Shauna A, James M (1972) Chemical basis of the sulfo–phospho-vanillin reaction for estimating total serum lipids. Clin Chem 18:199–202
Lagergren S, Svenska BK (1898) Zur theorie der sogenannten adsorption geloester stoffe. Veternskapsakad Handlingar 24:1–39
Ho YS, Mckay G (1998) Kinetic models for the sorption of dye from aqueous solution by wood. Transl chem E 76B:183–191
Box GEP, Hunter WG, Hunter JS (1978) Statistics for experimenters. Wiley, New York
Li Y, Cui FJ, Liu ZQ, Xu ZQ, Xu YY, Zhao H (2007) Improvement of xylanase production by Penicillium oxalicum ZH-30 using response surface methodology. Enzyme Microb Technol 40:1381–1388
Liu JZ, Weng LP, Zhang QL, Xu H, Ji LN (2003) Optimization of glucose oxidase production by Aspergillus niger in a bench top bioreactor using response surface methodology. World J Microbiol Biotechnol 19:317–323
Li Y, Lu J (2005) Characterization of the enzymatic degradation of arabinoxylans in grist containing wheat malt using response surface methodology. J Am Soc Brew Chem 63:171–176
Ghosh PK, Saxena RK, Gupta R, Yadav RP, Sheba D (1996) Microbial lipases: production and applications. Sci Prog 79(2):119–157
Gaur R, Gupta A, Khare SK (2008) Purification and characterization of lipase from solvent tolerant Pseudomonas aeruginosa PseA. Process Biochem 43:1040–1046
Chartrain M, Katz L, Marcin C, Thien M, Smith S, Fisher E et al (1993) Purification and characterization of a novel bioconverting lipase from Pseudomonas aeruginosa MB 5001. Enzyme Microb Technol 15:575–580
Ramani K, John Kennedy L, Ramakrishnan M, Sekaran G (2010) Purification, characterization and application of acidic lipase from Pseudomonas gessardii using beef tallow as a substrate for fats and oil hydrolysis. Process Biochem 45:1683–1691
Ramani K, Chockalingam Evvie, Sekaran G (2010) Production of a novel extracellular acidic lipase from Pseudomonas gessardii using slaughterhouse waste as a substrate. J Ind Microbiol Biotechnol 37:531–535
Ramani K, Sekaran G (2012) Production of lipase from Pseudomonas gessardii using blood tissue lipid and thereof for the hydrolysis of blood cholesterol and triglycerides and lysis of red blood cells. Bioprocess Biosyst Eng (article in press). doi:10.1007/s00449-011-0673-1
Jinwal UK, Roy U, Chowdhury AR, Bhaduri AP, Roy PK (2003) Purification and characterization of an alkaline lipase from a newly isolated Pseudomonas mendocina PK-12CS and chemoselective hydrolysis of fatty acid ester. Bioorg Med Chem 11:1041–1046
Karadzic I, Masui A, Zivkovic LI, Fujiwara N (2006) Purification and characterization of an alkaline lipase from Pseudomonas aeruginosa isolated from putrid mineral cutting oil as component of metal working fluid. J Biosci Bioeng 102:82–89
Voet D, Voet JG, Pratt CW (1999) Fundamentals of biochemistry. Wiley, New York
Wolff AM, Showell MS, Venegas MG, Barnett BL, Wertz WC (1996) Laundry performance of subtilisin protease. In: Bott RC (ed) Subtilisin enzymes: practical protein engineering. Plenum Press, New York, pp 6113–6120
Singh M, Banerjee UC (2007) Enantioselective transesterification of (RS)-1-chloro-3-(3,4-difluorophenoxy)-2-propanol using Pseudomonas aeruginosa lipases. Tetrahedron Asymmetry 18:2079–2085
Guncheva M, Zhiryakova D, Radchenkova N, Kambourova M (2007) Effect of nonionic detergents on the activity of a thermostable lipase from Bacillus stearothermophilus MC7. J Mol Catal B Enzym 49:88–91
Liebeton K, Zacharias A, Jaeger KE (2001) Disulfide bond in Pseudomonas aeruginosa lipase stabilizes the structure but is not required for interaction with its Foldase. J Bacteriol 183(2):597–603
Gupta M, Batra R, Tyagi R, Sharma A (1997) Polarity index: the guiding solvent parameter for enzyme stability in aqueous-organic cosolvent mixtures. Biotechnol Prog 13:284–288
Klibanov AM (2001) Improving enzymes by using them in organic solvents. Nature 409:241–246
Yadav RP, Agarwal P, Upadhay SN (2000) Microbial lipases: tools for drug discovery. J Sci Ind Res 59:977–987
Sharma R, Chisti Y, Chand BU (2001) Production, purification, characterization and applications of lipases. Biotechnol Adv 19:627–662
Surinenaite B, Bendikiene V, Juodka B (2009) The hydrolytic activity of Pseudomonas mendocina 3121–1 lipase. A kinetic study. Biologija 55:71–79
Ramani K, John Kennedy L, Vidya C, Boopathy R, Sekaran G (2010) Immobilization of acidic lipase derived from Pseudomonas gessardii onto mesoporous activated carbon for the hydrolysis of olive oil. J Mol Catal B Enzym 62:59–66
Wu XY, Jaaskelainen S, Linko Y (1996) An investigation of crude lipase for hydrolysis, esterification and transesterification. Enzyme Microb Technol 19:226–231
Acknowledgments
Authors Dr. K. Ramani and P. Saranya are thankful to Council of Scientific and Industrial Research (CSIR) and the Central Leather Research Institute (CLRI), India, for awarding senior research fellowship and providing all the facilities needed to carry out this work.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ramani, K., Saranya, P., Jain, S.C. et al. Lipase from marine strain using cooked sunflower oil waste: production optimization and application for hydrolysis and thermodynamic studies. Bioprocess Biosyst Eng 36, 301–315 (2013). https://doi.org/10.1007/s00449-012-0785-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-012-0785-2