Abstract
Fish meal has been used as an additional nitrogen source and fish oil as inducer for the growth and production of lipase from Cryptococcus sp. MTCC 5455. A response surface design illustrated that the optimum factors influencing lipase production were fish meal, 1.5 %, w/v, Na2HPO4, 0.2 %, w/v, yeast extract, 0.25 %, w/v and sardine oil, 2.0 %, w/v with an activity of 71.23 U/mL at 96 h and 25 °C, which was 48.39 % higher than the conventional one-factor-at-a-time method. The crude concentrated enzyme hydrolyzed polyurethane (PUR) efficiently and hydrolysis was 94 % at 30 °C and 96 h. The products, diethylene glycol and adipic acid were quantified by HPLC and scanning electron microscopic studies of the degraded polymer showed significant increase in size of the holes from 24 to 72 h of incubation. Hydrolysis of PUR within 96 h makes the lipase novel for disposal of PUR and provides an innovative solution to the problems created by plastic wastes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Fish processing generates considerable quantity of waste and nearly 63.6 million metric tonnes (MMT) of waste is generated globally, out of which 2.8 MMT are from India (Rai et al. 2010). Recovery of components with potential biological activities provides a means for value addition to the fish processing waste. These wastes are rich in protein and oil, which can be processed into useful products like fish protein concentrate, fish meal, fish silage, animal feed, etc.
Fish meal one of the main products obtained from fish waste, is relatively a dry product composed mainly of protein (70 %), minerals (10 %), fat (9 %) and water (8 %) (Pérez-Gálvez et al. 2012). Fish protein hydrolysate has been used as one of the ingredients in the media for the growth of bacteria such as Bacillus subtilis (Ellouz et al. 2001), Enterococcus durans (Ramakrishnan et al. 2013) and Bacillus cereus BG1 (Sellami-Kamoun et al. 2011) and fungi, including Rhizopus oryzae (Ghorbel et al. 2005) and Aspergillus oryzae (Garcia-Gomez et al. 2009). However, the reports on production of enzymes using fish meal, as a source of protein (Awan et al. 2010) and fish oil, as inducers (Ikemoto and Ota 1996; Kamini et al. 2000) are very much limited.
Lipases (triacylglycerol acyl hydrolases, E.C. 3.1.1.3) are an important group of biotechnologically relevant enzymes as they catalyze several reactions: hydrolysis, esterification, alcoholysis, aminolysis, peroxidations, epoxidations and interesterification (Contesini et al. 2010). The apparent promiscuity of these enzymes make lipases useful in numerous industrial processes, such as the synthesis of food ingredients (Dhake et al. 2011), their use as additives to detergents (Thirunavukarasu et al. 2008) and to obtain enantiopure drugs (Lou et al. 2006) and other refined products. Among microbial lipases, yeast lipases have received less attention and few reports are available on lipase producing yeasts such as Yarrowia lipolytica (Pignede et al. 2000), Candida antarctica (Lou et al. 2006) and C. rugosa (Herbst et al. 2012) and only limited research has been directed towards the lipase of Cryptococcus sp. (Chen et al. 1997). The Cryptococcus sp. MTCC 5455 (earlier as Cryptococcus sp. S-2) has been reported to produce an extracellular lipase in liquid medium with triolein as an inducer (Kamini et al. 2000). This enzyme could be effectively used in the production of methyl esters, which are excellent substitutes for diesel fuel (Kamini and Iefuji 2001) and as an additive in detergent formulation (Thirunavukarasu et al. 2008). Furthermore, the purified enzyme could effectively degrade the high molecular weight compound polylactic acid (PLA) and other biodegradable plastics including polybutylene succinate (PBS) and polycaprolactone (PCL) (Masaki et al. 2005).
In the industrialization of fermentation processes, production costs are greatly influenced by the price of the media components, mainly protein sources. The utilization of locally available natural nutritive resources like addition of fish meal to the production media could reduce the cost of the medium, thereby making the process economically attractive, viable and green. Fish meal contains the essential substances like carbon, nitrogen and minerals that are essential for the growth of microorganisms (Esakkiraj et al. 2010). Fish peptones and fish protein hydrolysate have been used as nitrogen source for the production of various metabolites, such as protease (Ellouz et al. 2001; Garcia-Gomez et al. 2009; Sellami-Kamoun et al. 2011), lipase (Souissi et al. 2009) and β-galactosidase (Awan et al. 2010). Similarly, sardine oil has been reported as a good inducer for production of lipase from Geotrichum sp. FO 274A (Ikemoto and Ota 1996) and Cryptococcus sp. (Kamini et al. 2000). In this study, central composite rotatable design (CCRD), a tool of RSM has been used to determine the optimal conditions for production of an extracellular lipase from Cryptococcus sp. MTCC 5455 using yeast extract, fish meal, a fishery by-product used for the first time as an additional complex nitrogen source, phosphates and fish oil, an inducer, which were found to be optimum by the classical one-factor-at-a-time method. Furthermore, the lipase was concentrated by ultrafiltration and evaluated for the hydrolysis of polyurethane (PUR) in aqueous medium. Polyurethanes are widely used in various industries because of its high mechanical properties and thermal insulations. The global consumption of PUR has been reported as 13,650,000 tonnes in 2010, while it was 215,000 tonnes in India and the demand increased to about 350,000 tonnes in 2012. As the demand increases, their disposal in landfills and incinerators causes significant environmental and resource depletion problems (Gautam et al. 2007a). The thermal degradation of PUR results in the production of toxic products like hydrogen cyanide (HCN) and nitrogen oxides (NOx), which have negative impact on environment as reported by Risholm-Sundman and Vestin (2005). Few microorganisms are known to degrade polyurethane (Crabbe et al. 1994; Nakajima-Kambe et al. 1995; Rowe and Howard 2002; Gautam et al. 2007b) and the reports on lipase mediated PUR degradation are very much limited (Kim and Kim 1998). Enzymatic degradation is suggested to be an alternative to the conventional process as the hydrolytic products, organic acids and polyols could be recovered and used for chemical recycling of polymers apart from solid waste management. Therefore, an attempt has been made to evaluate the degradation of PUR by Cryptococcus sp. lipase to make the process viable. Moreover, fish meal and fish oil have been explored for the growth and production of lipase in order to reduce the material cost of the medium.
Materials and methods
Materials
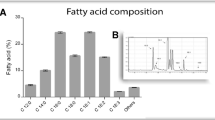
All the chemicals used in the present study were of analytical reagent (AR) grade and purchased from Hi-Media Limited and SD Fine Chemicals Limited, Mumbai, India. Fish meal and fish oils (cod-liver oil, shark oil and sardine oil) were obtained as gifts from Raj Fisheries Limited, Mangalore, India, respectively. The fish meal was analysed for the contents of protein, lipids, ash and moisture as per the protocols described by Ellouz et al. (2001). The cod-liver oil, shark oil and sardine oil contained 21, 23 and 39 % saturated fatty acids, 41, 25 and 26 % monounsaturated fatty acids and 31, 48 and 33 % polyunsaturated fatty acids, respectively as given by Raj Fisheries Limited. The other ingredients of the media, namely, sesame oil, sunflower oil, soybean oil and olive oil were purchased from local markets in Chennai, India. The poly(diethylene glycol adipate) was kindly supplied by Suzuki Motor Co., Hamamatsu, Japan and 2,4-tolylene diisocyanate was purchased from Wako Pure Chemical Co., Tokyo, Japan, respectively.
Synthesis of polyurethane (PUR)
The polyester polyurethane used in this study was synthesized by reacting poly(diethylene glycol adipate)s with 2,4-tolylene diisocyanate (TDI) under anhydrous condition as described by Nakajima-Kambe et al. (1995).
Inoculum and culture conditions
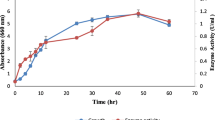
The organism was isolated from air and identified as Cryptococcus sp. MTCC 5455 (earlier reported as Cryptococcus sp.S-2) (Kamini et al. 2000) and maintained on potato dextrose agar (PDA) slants at 4 °C. It was grown in the YM medium (3 g/L yeast extract, 5 g/L malt extract, 5 g/L peptone and 10 g/L dextrose, pH 6.2 ± 0.2) for 40 h at 25 °C (5.4–5.8 × 108 cells/mL) which was used as the inoculum. Lipase production was carried out in 250 mL Erlenmeyer flasks containing 40 mL of the sterile production medium (pH 5.6) and the medium was inoculated with 1 % (v/v) of the inoculum and grown for 120 h on a rotary shaker at 25 °C and 100 rpm. At 24 h intervals, 3 mL of the samples were withdrawn, centrifuged at 5867 g for 10 min and the supernatant was assayed for the enzyme activity.
Optimization studies
Lipase production was initially carried out in the optimized production medium of Kamini et al. (2000) with the following composition (per liter, w/v): yeast extract, 10.0 g; lactose, 5.0 g; KH2PO4, 10.0 g; MgSO4.7H2O, 1.0 g and triolein, 10.0 g. To develop a low-cost medium, optimization studies were investigated by the classical one-factor-at-a-time method using yeast extract (0.5–1.5 %, w/v), fish meal (0.5–2.0 %, w/v), lactose (0.5 %, w/v), MgSO4.7H2O (0.01 %, w/v), various inorganic nitrogen sources (NH4NO3, (NH4)2SO4, NH4H2PO4 and NaNO3, 0.5 % w/v), phosphates (KH2PO4, K2HPO4, NaH2PO4 and Na2HPO4, 0.2–1.0 %, w/v) and inducers (triolein, olive oil, cod-liver oil, shark oil, sardine oil, soybean oil, sesame oil and sunflower oil, 1.0–2.0 %, w/v) for the production of lipase. The C/N ratio of the basal medium and optimized medium was determined using automated elemental analyzer (Vario MICRO, Elementar Analysensysteme GmbH, Hanau, Germany).
Enzyme assay
The lipase activity was estimated by the spectrophotometric method using p-nitrophenyl laurate (p-NPL) as a substrate (Isobe et al. 1988). One unit of lipase activity was defined as the amount of enzyme that liberates 1 μmol of p-nitrophenol per minute under the standard assay conditions.
Factorial design
A statistical approach was employed to study the interaction of critical variables for optimum lipase production by RSM, which identifies possible interactions, high order effects and optimum operational fermentation conditions. On the basis of the results obtained by the classical method, a five-level four-factorial central composite rotatable design (CCRD) was employed in this study with an alpha value of ±1.414 for the selected factors: A (fish meal), B (sardine oil), C (Na2HPO4) and D (yeast extract). The relationship between the variation of the response, Yc (lipase activity, U/mL) and the variation of factors A-D is represented by a second-order mathematical model using the equation \( \begin{array}{c}\hfill Yc={\beta}_0+{\beta}_1{A}_1+{\beta}_2{B}_2+{\beta}_3{C}_3+{\beta}_4{D}_4\left(\mathrm{intercept}\kern0.5em \mathrm{and}\kern0.5em \mathrm{main}\kern0.5em \mathrm{effects}\right)\hfill \\ {}\hfill +{\beta}_{11}{A_1}^2+{\beta}_{22}{B_2}^2+{\beta}_{33}{C_3}^2+{\beta}_{44}{D_4}^2\left(\mathrm{interactions}\right)\hfill \\ {}\hfill +{\beta}_{12}{A}_1{B}_2+{\beta}_{13}{A}_1{C}_3+{\beta}_{14}{A}_1{D}_4+{\beta}_{23}{B}_2{C}_3+{\beta}_{24}{B}_2{D}_4+{\beta}_{34}{C}_3{D}_4\left(\mathrm{quadratic}\kern0.5em \mathrm{effects}\right)\hfill \end{array} \) where, Yc was the response calculated by the model, β 0 represented the regression coefficient at the center; A 1 , B 2 , C 3 , D 4 were the coded variables corresponding to factors A, B, C and D; β 1, β 2, β 3, β 4, linear coefficients; β 11, β 22, β 33, β 44, squared coefficients and β 12, β 13, β 14 , β 23, β 24, β 34 were the interaction coefficients. The combinations of CCRD were allocated in three blocks and each block had 10 runs. The first two blocks each had eight factorial points and two center points. The last block had eight axial points and two center points. Thus, in total, the experimental setup consisted of 30 trials and the value of the dependent response was the mean of triplicates. Analysis of variance (ANOVA), regression analysis and three dimensional response surface curves were plotted by the ‘Design Expert’ software package (Version 7.0.3, Stat-Ease Inc., Minneapolis, USA) to study the interaction among the factors and production of lipase.
Hydrolysis of polyurethane by Cryptococcus sp. lipase
The concentrated enzyme (specific activity, 11.90 U/mg of protein) obtained by ultrafiltration was used for the hydrolysis of polyurethane. Hydrolysis of PUR was carried out in series of 50 mL screw capped Erlenmeyer flasks containing 50 mg of cube-shaped PUR, 5 mL of 0.1 M phosphate buffer (pH 7.0) and 1000 U of lipase and incubated at 30 °C for 120 h with shaking at 120 rpm. A control was run simultaneously with the concentrated supernatant obtained from the medium without inducer (sardine oil) in order to evaluate the PUR hydrolysis. The degradation was monitored at 24 h intervals by measuring the weight of PUR before and after incubation. The PUR cubes were retrieved from the reaction mixture by filtering through nylon net filters with pore size of 10 μm. The PUR cubes were washed twice with distilled water, air dried and weighed. The effect of lipase concentration (500–2000 U) on degradation of PUR was also studied to achieve maximum hydrolysis and the hydrolyzed products were quantitatively determined using high performance liquid chromatography (HPLC) as per the method described by Nakajima-Kambe et al. (1997). For HPLC analysis, 900 μl of the reaction mixture (after removing the PUR cubes) was mixed with 100 μl of 0.1 N H2SO4 and incubated at room temperature for 10 min. The reaction mixture was centrifuged at 5867 g for 5 min and 20 μl of the supernatant was injected into a HPLC system (model 2695, Waters Co., MA, USA), equipped with a HPX-87(H) column (7.8 mm diameter × 30 cm, Bio-Rad Laboratories Inc., CA, USA) and a refractive index (RI) detector (model 2414, Waters Co.). The mobile phase was 5 mM H2SO4 and the flow rate was 0.7 mL/min. For the determination of the metabolites derived from polyisocyanate segments of the PUR, the supernatant was alkalized to pH 13.0 with 1 N NaOH and incubated at 121 °C for 20 min. The alkaline-hydrolyzed PUR metabolite was extracted with ethyl acetate and 10 μl of the extract was injected into the HPLC system set with RP-18 column (5 μm) Merck KgaA, Darmstadt, Germany. The mobile phase consisted of acetonitrile and alkaline water (pH 9.0) in the ratio of 37.5: 62.5 and the flow rate was 0.7 mL/min. The changes in surface texture of PUR during the reaction were also observed at various time intervals by scanning electron microscope (SEM). PUR cubes retrieved from the 50 ml screw capped flasks were washed thoroughly with distilled water, air-dried and placed on the adhesive carbon tapes mounted on SEM specimen stub. The samples were coated with gold in argon medium and surface was scanned using JEOL JM-5600 electron microscopic unit at 20 kV accelerating voltage.
Results and discussion
The production of lipase from Cryptococcus sp. MTCC 5455 in submerged fermentation was reported earlier by Kamini et al. (2000) with triolein as an inducer, which contributes to 96 % of the medium cost, followed by yeast extract. Therefore, attempts were made to make the process feasible by using inexpensive fishery by-products, such as fish oil to replace triolein and fish meal to substitute yeast extract completely or partially. The fish meal (on dry weight basis) contained 59 ± 2.8 % protein, 10 ± 0.8 % lipid, 9 ± 0.3 % ash and 7.5 ± 0.3 % moisture. By conventional one factor-at-a-time approach, an optimal lipase activity of 48 U/mL was obtained at 96 h with a combination of 1.0 % FM (w/v) and 0.5 % yeast extract (w/v), thereby reducing the yeast extract to 50 % and by replacing triolein with 1.5 % sardine oil (w/v), respectively, in a medium supplemented with lactose, 0.5 % (w/v); MgSO4.7H2O, 0.1 % (w/v) and Na2HPO4, 0.4 % (w/v) (data not shown). This lipase activity was comparatively higher than the lipase activities of Rhizopus oryzae (15 U/mL), (Ghorbel et al. 2005) and Staphylococcus epidermidis CMST Pi 2 (14.2 U/mL) (Esakkiraj et al. 2010) in a medium supplemented with 4 % sardinella fish powder and 2.5 % defatted fish meal, respectively. Among fish oils, sardine oil was found to be an ideal inducer (48 U/mL) for production of lipase from Cryptococcus sp., followed by soybean oil (39.5 U/mL), olive oil (38 U/mL) and sesame oil (37.6 U/mL). However, the reported lipase activities were low in Bacillus licheniformis MTCC 6824 with sardine (13.6 U/mL) and cod liver oil (7.53 U/ mL) as inducers (Chakraborty and Raj 2008). In order to determine the interaction effects among the variables and to depict the net effect of the medium constituents for enhanced production of lipase, statistical optimization of the medium components was carried out by CCRD using sardine oil, fish meal, yeast extract and Na2HPO4.
CCRD model fitting and ANOVA
The coded and actual values of the variables are given in Table 1. The levels of the factors were chosen on the basis of the results obtained from the conventional one-factor-at-a-time approach. The experiments were performed to obtain a quadratic model with four independent variables as shown in Table 1. The quadratic polynomial model was seen to be highly significant to represent the actual relationship between the response (lipase activity) and the significant variables, as the observed values were found to correlate with the predicted values obtained from the model fitting technique using the software Design-Expert version 7.0.3. The variation in lipase activities from 11.46 to 71.23 U/mL indicated that the interactions among the factors played a more significant role than the effect of individual factors alone and emphasized the importance of medium optimization for higher lipase production. The ANOVA for the quadratic regression model (Table 2) showed that the model was highly significant, with an F value of 23.58, as is evident from Fisher’s F test along with a very low probability value (P model > F = 0.0001). At the same time, a relatively lower coefficient of variation (CV = 9.21 %) indicated a better precision and reliability of the experiments carried out. The determination coefficient (R 2) of the model was 0.9621, explaining 96.21 % of the variability in the response could be accounted to the independent variables and only 3.79 % of the total variation was not explained by the model.
Analysis of the design showed a high degree of fitting between predicted and experimental data, which indicated that the model suitably represented the real relationship among the selected factors. The insignificant lack of fit test also indicated that the model suitably represented the experimental data and the final predictive equation was as follows:
From this model, it was clear that three linear coefficients, two quadratic coefficients and three cross product coefficients were highly significant (P < 0.05, Table 2) and among the four variables, fish meal, sardine oil and yeast extract were the most significant for lipase production, while Na2HPO4 had less significance on production of lipase. The high F value of 196.40, in addition to a least probability value (<0.0001) for fish meal, indicated that fish meal played a very prominent role in lipase production, when compared to other variables. The cumulative effect of sardine oil and fish meal on lipase production in a medium containing 0.4 % (w/v) Na2HPO4 and 0.5 % (w/v) yeast extract is shown in the response surface plot of Fig. 1. The lipase activity was 55 U/mL, when the production medium contained 1.5 % (w/v) fish meal and 2.0 % (w/v) sardine oil, whereas decreasing the concentrations of fish meal and sardine oil reduced the lipase production levels, confirming that fish meal and sardine oil had significant influence on lipase production. Similar results were reported for the production of lipase from Rhizopus oryzae (Ghorbel et al. 2005), S. simulans (Souissi et al. 2009) and S. epidermidis CMST Pi 2 (Esakkiraj et al. 2010) using fish meal as an additional nitrogen source. To the best of our knowledge, vegetable oils or synthetic triglycerides (triolein and tributyrin) are used for inducing lipase production and the reports on using fish oils as inducer are very much limited (Chakraborty and Raj 2008; Esakkiraj et al. 2010). Moreover, this is the first study showing the highest lipase activity using sardine oil as an inducer along with fish meal as an additional complex nitrogen source, because both are renewable fishery by-products and cost-effective for production of lipase at an industrial scale. The material cost for the production medium of Kamini et al. (2000) was 86.20 ± 1.70 USD/l, which was comparatively higher than the cost-effective medium (0.64 ± 0.15 USD/l) including the cost of inoculum medium.
The interactive effect of Na2HPO4 and fish meal on the production of lipase is shown in Fig. 2. Maximum lipase activity of 62 U/mL was obtained with low concentration of Na2HPO4 (0.2 %, w/v) and at 1.50 % (w/v) fish meal concentration. The lipase activity increased with increasing concentrations of fish meal from 0.5 to 1.5 % (w/v), while the activity was not increased significantly with increasing concentrations of Na2HPO4 from 0.2 to 0.6 % (w/v). Similar results were reported for production of lipase from Pseudomonas fluorescens by Makhzoum et al. (1995) with optimal activity obtained at low concentration (1 mM) of phosphate. The cumulative effect of yeast extract and fish meal showed that the lipase activity was optimum (55.5 U/mL), when the media contained 0.25 % (w/v) of yeast extract and 1.5 % (w/v) of fish meal. Figure 3 depicts the interactive effect of yeast extract and sardine oil with an optimal lipase activity at 0.25 % (w/v) yeast extract and 2.0 % (w/v) sardine oil concentrations. The lipase activity increased with increasing concentrations of sardine oil from 1 to 2.0 % (w/v), whereas increase in concentration of yeast extract from 0.25 to 0.75 % had less influence on lipase activity. The design illustrated that the optimum values of the factors influencing the lipase production were 1.5 % (w/v) fish meal, 0.2 % (w/v) Na2HPO4, 0.25 % (w/v) yeast extract and 2.0 % (w/v) sardine oil, with an activity of 71.23 U/mL at 96 h and 25 °C, which was 48.39 % higher than the activity obtained by the conventional one-factor-at-a-time method. This could be due to the increase in C/N ratio from 4.91 (optimized medium of conventional one-factor-at-a-time method) to 5.63 (statistically optimized medium), while it was 4.52 for the production medium of Kamini et al. (2000). Similarly, the production of lipase from Pseudomonas aeruginosa was increased by three fold, when the C/N ratio was increased from 1.66 to 4.13 and there was no further increase in lipase production with increase in C/N ratio to 8.26 (Chartrain et al. 1993). Rao et al. (1993) reported an optimal lipase production at a C/N ratio of 6.0–6.5 for Candida rugosa, in a medium containing 0.5 % urea (w/v), 1.5 % maltose (w/v) and 7.5 % oil (w/v), while further increase in concentration of urea and maltose did not increase the lipase activity. Ghribi et al. (2009) also reported a decline in lipase production, when the C/N ratio was increased above 6.0 for lipase production from Staphylococcus xylosus. In the present study, the supplementation of yeast extract was reduced by 75 % compared to the production medium of Kamini et al. (2000), where 1 % yeast extract was used for the production of lipase with 1 % triolein as inducer. The maximal lipase activity (71.23 U/mL) observed under the optimized conditions was not depicted in response surface plots. This could be due to the fact that response surface plots were drawn by imposing constant values (i.e., the central points of the interval taken into consideration) to two of the independent variables of the factorial design. The lipase produced from Cryptococcus sp. was also evaluated for hydrolysis of PUR cubes.
Hydrolysis of PUR
The concentrated enzyme showed maximum activity at pH 7.0 to 8.0 and at 37 °C and was stable between pH 5.0 and 10.0 and at temperatures up to 60 °C (data not shown). The enzyme degraded the PUR efficiently and an optimal degradation of 94 % was obtained at 30 °C and 96 h with 1500 U of lipase (Fig. 4), while the degradation was 23 and 67 % using 500 and 1000 U of enzyme concentration, respectively and the degradation was not significantly increased upon increasing the enzyme concentration to 2000 U (95.2 %). On the other hand, the degradation was relatively small and moderate after 8 and 12 days of incubation with Candida cylindracea lipase (Kim and Kim 1998) and Pseudomonas chlororaphis esterase (Gautam et al. 2007b), respectively. The degradation of PUR by Cryptococcus sp. lipase was linear from 24 to 72 h with the production of diethylene glycol (DEG) and adipic acid (AA) and the total amounts of degraded products were almost equal to the theoretical yield of degraded products of PUR, while DEG or AA were not detected in the control. The enzyme could also be used for degradation of any commercial polyester type polyurethane waste, because the polyester polyol (poly(diethylene glycol adipate)s) used in this study is the same material, which is used for production of commercial polyurethane. However, the realistic conditions need to be standardized in future for PUR removal and remedial processes under in-vitro conditions.
The HPLC analysis of the degradation products, DEG and AA are shown in Fig. 5a and 2,4-diaminotoluene in Fig. 5b. The presence of 2,4-diaminotoluene obtained after the alkaline treatment indicated that the reaction mixture contained an unknown water soluble metabolite derived from polyisocyanate segment of PUR. SEM studies also supported the degradation of polyester PUR by the yeast lipase. SEM photomicrographs of the control (undegraded) and degraded PUR cubes at 24, 48 and 72 h are shown in Fig. 6. The surface of the PUR cubes showed small holes uniformly at 24 h followed by significant increase in size of the holes from 48 to 72 h clearly indicated the degradation of PUR by Cryptococcus sp. lipase. These results indicated that the Cryptococcus sp. lipase could be effectively used in the transformation of PUR into water soluble compounds and the hydrolytic products could be recovered and used for chemical recycling of polymers.
Conclusion
The utilization of fish meal, a complex nitrogen source could reduce the supplementation of yeast extract to 75 % and sardine oil, by replacing triolein, led to a reduction in the overall cost of production medium by 90–96 % of the total medium cost, thereby making the process convincingly attractive and viable. Furthermore, the lipase could be effectively utilized in the hydrolysis of PUR, since environmental pollution by plastic wastes has become a serious issue recently. Therefore, degradation of PUR by Cryptococcus sp. lipase makes the process viable and provides an innovative solution for the disposal of PUR.
References
Awan MS, Khan SA, Rehman ZU, Saleem A, Rana SM, Rajoka MI (2010) Influence of nitrogen sources on production of β-galactosidase by Aspergillus niger. Afr J Biotechnol 9:2918–2922
Chakraborty K, Raj RP (2008) An extra-cellular alkaline metallolipase from Bacillus licheniformis MTCC 6824: purification and biochemical characterization. Food Chem 109:727–736
Chartrain M, Marcin C, Katz L, Salmon P, Brix T, Buckland B, Greasham R (1993) Enhancement of lipase production during fed-batch cultivation of Pseudomonas aeruginosa MB 5001. J Ferment Bioeng 76:487–492
Chen LC, Pirofski LA, Casadevall A (1997) Extracellular proteins of Cryptococcus neoformans and host antibody response. Infect Immun 65:2599–2605
Contesini FJ, Lopes DB, Macedo GA, Nascimento MD, Carvalho PD (2010) Aspergillus sp lipase: potential biocatalyst for industrial use. J Mol Catal B Enzym 67:163–171
Crabbe JR, Campbell JR, Thompson L, Walz SL, Schultz WW (1994) Biodegradation of a colloidal ester-based polyurethane by soil fungi. Int Biodeterior Biodegradation 33:103–113
Dhake KP, Krishna MD, Yogesh PP, Rekha SS, Bhalchandra MB (2011) Improved activity and stability of Rhizopus oryzae lipase via immobilization for citronellol ester synthesis in supercritical carbon dioxide. J Biotechnol 156:46–51
Ellouz Y, Bayoudh A, Kammoun S, Gharsallah N, Nasri M (2001) Production of protease by Bacillus subtilis grown on sardinelle heads and viscera flour. Bioresour Technol 80:49–51
Esakkiraj P, Dhas GAJ, Palavesam A, Immanuel G (2010) Media preparation using tuna-processing wastes for improved lipase production by shrimp gut isolate Staphylococcus epidermidis CMST Pi 2. Appl Biochem Biotechnol 160:1254–1265
Garcia-Gomez MJ, Huerta-Ochoa S, Loera-Corral O, Prado-Barragan LA (2009) Advantages of a proteolytic extract by Aspergillus oryzae from fish flour over a commercial proteolytic preparation. Food Chem 112:604–608
Gautam R, Bassi AS, Yanful EK (2007a) A review of biodegradation of synthetic plastic and foams. Appl Biochem Biotechnol 141:85–108
Gautam R, Bassi AS, Yanful EK, Cullen E (2007b) Biodegradation of automotive waste polyester polyurethane foam using Pseudomonas chlororaphis ATCC55729. Int Biodeterior Biodegradation 60:245–249
Ghorbel S, Souissi N, Triki-Ellouz Y, Dufosse L, Guerard F, Nasri M (2005) Preparation and testing of Sardinella protein hydrolysates as nitrogen source for extracellular lipase production by Rhizopus oryzae. World J Microbiol Biotechnol 21:33–38
Ghribi D, Sayari A, Gargouri Y, Bezzine S (2009) Improvement of Staphylococcus xylosus lipase production through optimization of the culture conditions. Eur J Lipid Sci Technol 111:967–971
Herbst D, Peper S, Niemeyer B (2012) Enzyme catalysis in organic solvents: influence of water content, solvent composition and temperature on Candida rugosa lipase catalyzed transesterification. J Biotechnol 162:398–403
Ikemoto M, Ota Y (1996) Production of two types of non-specific lipases by Geotrichum sp FO 274A: a fish oil-assimilating strain. J Gen Appl Microbiol 42:371–379
Isobe K, Akiba T, Yamaguchi S (1988) Crystallization and characterization of lipase from Penicillium cyclopium. Agric Biol Chem 52:41–47
Kamini NR, Iefuji H (2001) Lipase catalyzed methanolysis of vegetable oils in aqueous medium by Cryptococcus spp. S-2. Process Biochem 37:405–410
Kamini NR, Fujii T, Kurosu T, Iefuji H (2000) Production, purification and characterization of an extracellular lipase from the yeast, Cryptococcus sp S-2. Process Biochem 36:317–324
Kim YD, Kim SC (1998) Effect of chemical structure on the biodegradation of polyurethanes under composting conditions. Polymer Degrad Stab 62:343–352
Lou WY, Zong MH, Liu YY, Wang JF (2006) Efficient enantioselective hydrolysis of D, L-phenylglycine methyl ester catalyzed by immobilized Candida antarctica lipase B in ionic liquid containing systems. J Biotechnol 125:64–74
Makhzoum A, Knapp JS, Owusu RK (1995) Factors affecting growth and extracellular lipase production by Pseudomonas fluorescens 2D. Food Microbiol 12:277–290
Masaki K, Kamini NR, Ikeda H, Iefuji H (2005) Cutinase-like enzyme from the yeast Cryptococcus sp strain S-2 hydrolyzes polylactic acid and other biodegradable plastics. Appl Environ Microbiol 71:7548–7550
Nakajima-Kambe T, Onuma F, Kimpara N, Nakahara T (1995) Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen-source. FEMS Microbiol Lett 129:39–42
Nakajima-Kambe T, Onuma F, Akutsu Y, Nakahara T (1997) Determination of the polyester polyurethane breakdown products and distribution of the polyurethane degrading enzyme of Comamonas acidovorans stain TB-35. J Ferment Bioeng 83:456–460
Pérez-Gálvez R, Guadix A, Almécija MC, Guadix EM, Bergé JP (2012) Response surface modeling of the multiphase juice composition from the compaction of sardine discards. Food Bioprocess Technol 5:2172–2182
Pignede G, Wang HJ, Fudalej F, Gaillardin C, Seman M, Nicaud JM (2000) Characterization of an extracellular lipase encoded by LIP2 in Yarrowia lipolytica. J Bacteriol 182:2802–2810
Rai AK, Swapna HC, Bhaskar N, Halami PM, Sachindra NM (2010) Effect of fermentation ensilaging on recovery of oil from fresh water fish viscera. Enzyme Microb Technol 46:9–13
Ramakrishnan V, Goveas LC, Halami PM, Narayan B (2013) Kinetic modeling, production and characterization of an acidic lipase produced by Enterococcus durans NCIM5427 from fish waste. J Food Sci Technol. doi:10.1007/s13197-013-1141-5
Rao PV, Jayaraman K, Lakshmanan CM (1993) Production of lipase by Candida rugosa in solid state fermentation. 2: medium optimization and effect of aeration. Process Biochem 28:391–395
Risholm-Sundman M, Vestin E (2005) Emissions during combustion of particleboard and glued veneer. Holz Als Roh-Und Werkstoff 63:179–185
Rowe L, Howard GT (2002) Growth of Bacillus subtilis on polyurethane and the purification and characterization of a polyurethanase-lipase enzyme. Int Biodeterior Biodegradation 50:33–40
Sellami-Kamoun A, Ghorbel-Frikha B, Haddar A, Nasri M (2011) Enhanced Bacillus cereus BG1 protease production by the use of sardinelle (Sardinella aurita) powder. Ann Microbiol 61:273–280
Souissi N, Bougatef A, Triki-ellouz Y, Nasri M (2009) Production of lipase and biomass by Staphylococcus simulans grown on sardinella (Sardinella aurita) hydrolysates and peptone. Afr J Biotechnol 8:451–457
Thirunavukarasu K, Edwinoliver NG, Anbarasan SD, Gowthaman MK, Iefuji H, Kamini NR (2008) Removal of triglyceride soil from fabrics by a novel lipase from Cryptococcus sp S-2. Process Biochem 43:701–706
Acknowledgments
The authors thank The Director, Central Leather Research Institute, Chennai, India for his kind permission to publish this work. We also acknowledge for the grants from Department of Biotechnology, Government of India and the financial assistance extended by Council of Scientific and Industrial Research, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Thirunavukarasu, K., Purushothaman, S., Gowthaman, M.K. et al. Utilization of fish meal and fish oil for production of Cryptococcus sp. MTCC 5455 lipase and hydrolysis of polyurethane thereof. J Food Sci Technol 52, 5772–5780 (2015). https://doi.org/10.1007/s13197-014-1697-8
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13197-014-1697-8