Abstract
Zinc ion plays essential roles in biological chemistry. Bacteria acquire Zn2+ from the environment, and cellular concentration levels are controlled by zinc homeostasis systems. In comparison with other homeostatic systems, the ZraSR two-component system was found to be more efficient in responding to exogenous zinc concentrations. To understand the dynamic response of the bacterium ZraSR two-component system with respect to exogenous zinc concentrations, the genetic circuit of the ZraSR system was integrated with a reporter protein. This study was helpful in the construction of an E. coli system that can display selective metal binding peptides on the surface of the cell in response to exogenous zinc. The engineered bacterial system for monitoring exogenous zinc was successfully employed to detect levels of zinc as low as 0.001 mM, which directly activates the expression of chimeric ompC t —zinc binding peptide gene to remove zinc by adsorbing a maximum of 163.6 μmol of zinc per gram of dry cell weight. These results indicate that the engineered bacterial strain developed in the present study can sense the specific heavy metal and activates a cell surface display system that acts to remove the metal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zinc ion is an essential metal for normal cell function, but can be toxic at elevated concentrations, a dual role it plays in all existing cells [1]. Almost all biological interactions between proteins and Zn2+ may be used to facilitate correct folding and to stabilize protein structure [2, 3]. Zn2+ plays a vital catalytic role in many proteins and accumulates at the same levels as other important elements, namely calcium and iron, in Escherichia coli [4]. However, zinc has an essential role in metabolic activity, and it is found to be highly cytotoxic when accumulated in excess. To overcome this problem, nature provides a tightly regulated homeostatic mechanism in all life forms to deal with abnormally low and high levels of zinc [5]. In E. coli, a number of chromosomal genes involved in zinc homeostasis systems have been identified. As a result, starvation and toxicity by zinc in cells leads to the transcription of a number of genes, which are regulated by zinc inducible promoters [6]. Among these genes, the zraR and zntR were found to respond better to zinc metal, through involvement in transcriptional regulation of zinc homeostasis genes.
ZraP is a 20.4 kDa membrane-associated protein that undergoes a specific Zn2+-induced cleavage to release a 12 kDa carboxy-terminal Zn2+-binding region into the periplasm that is involved in the acquisition of tolerance to high Zn2+ concentrations. The expression of zraP is regulated by ZraSR (previously called HydHG), a membrane-associated sensor kinase and a response regulator, which is involved in transcription regulation of zinc homeostasis genes. In response to high Zn2+ concentrations, ZraR and ZraS specifically activate zraP, which is divergently transcribed from zraSR, and also autogenously activate zraSR expression [7]. The zraSR appears to have a weak constitutive promoter, which ensures basal synthesis of the sensor and response regulator [8, 9]. Moreover, transcription activators of zraP expression have a σ54 dependent promoter located in the zraP–zraSR intergenic region where ZraR binds. An important property of this promoter is that transcription can be completely turned off in the absence of transcription activators [10].
The zntA–zntR system present in the bacterial cytoplasm, which encodes the zinc efflux protein and Zn2+ binding MerR-like transcriptional activator, respectively, leading to detoxification of zinc [11–13]. At higher concentrations of Zn2+, the system fails to efflux the metal and is unable to sense exogenous metal [14]. However, ZraSR, a two-component membrane associated sensor kinase system, senses exogenous zinc and responds preferentially to Zn2+ even at higher concentrations.
In this paper, we studied the dynamic characteristics of zraP gene expression by exogenous Zn2+ using quantitative real-time PCR (RT-PCR) and green fluorescence protein based reporter system. Then a simple zinc adsorption system which adsorbs exogenous Zn2+ on the bacterial surface was constructed (Fig. 1). For the display of zinc binding peptides on the surface of E. coli, OmpC was used as an anchoring motif and it was integrated with of the E. coli zraP promoter (P zraP ). Hence, this adsorption system is activated in responding to exogenous zinc and adsorbs zinc on its surface.
Engineered E. coli for sensing and removing heavy metals. Periplasmic metal sensing receptors sense Zn2+, which phosphorylate the histidine kinase domain and response regulator from the OmpR family of TCS. This activates the synthetic genetic circuit of ZraSR (HydHG) TCSs resulting in Zn2+ binding peptides displayed on the E. coli surface for bioremediation
Materials and methods
Bacterial strains and media
E. coli XL1-Blue was used as the host strain for recombinant DNA manipulation. Plasmids and bacterial strains used in this study are listed in Table 1. All recombinant E. coli were cultivated in Luria–Bertani (LB) medium (10 g/l bacto-tryptone, 5 g/l bacto-yeast extract and 5 g/l NaCl) and M9 minimal salts medium, unless otherwise stated, with glucose (0.4%) as a carbon source and supplements of 2 mM MgSO4, 0.1 mM CaCl2, and 1% thiamine HCl per ml supplemented with antibiotics (ampicillin, 100 mg/l) at 37 °C, with vigorous shaking (200 rpm).
Construction of plasmid
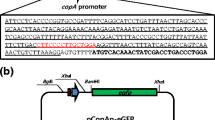
The genomic region containing the 237 bp zraP–zraS intergenic region was amplified from E. coli genomic DNA with the zraP_FBamHI and zraP_RSalI oligonucleotides (Table 2). Polymerase chain reaction (PCR) was performed with a MJ mini Personal Thermal Cycler (BioRad Laboratories, USA) using the Expand High Fidelity PCR System (Roche Molecular Biochemicals, Mannheim, Germany). The PCR product was digested with BamHI and SalI and was ligated with pUC19 to construct the pUHup1 plasmid [15]. The gfp gene encoding GFP was amplified from plasmid pPROBE-NT′ [16] and was ligated with pUHup1 using BamHI/KpnI restriction enzymes to construct pZGFP1 in which the GFP reporter protein was under the control of a zraP promoter (Fig. 2a).
a Plasmid pZGFP1 constructed to sense Zn2+ in bacterial cultures. b E. coli system effectively senses Zn2+ in bacterial cultures. The localization and expression of GFP under the control of the zraP promoter were analyzed by reflected fluorescence microscopy. E. coli carrying pZGFP1 sensed Zn2+ (presence of 1 mM ZnCl2). Magnification ×100
E. coli ompC was amplified from the genomic DNA of E. coli with designed oligonucleotides based on the reported genome sequences [17]. For expression of the zinc binding peptide (HYQHNTHHPSRW) on the cell surface [18], truncated ompC (ompC t ) genes encoding the 331 amino acids from the N-terminus were amplified using two complimentary pairs of oligonucleotides, shown in Table 2 [19, 20]. The PCR product was cloned into the pUHup1 plasmid using KpnI and BamHI to construct pZZ1056 in which the chimeric protein was under the control of a zraP promoter. These plasmids were transformed into chemically competent E. coli cells for further studies.
Expression monitoring of zraP gene
The transcriptional activities of the zraP gene in response to Zn2+ in E. coli cells harboring pZGFP1 were measured by quantitative RT-PCR. A single colony of E. coli harboring pZGFP1 was grown overnight in nutrient rich LB medium and minimal M9 medium was incubated at 37 °C in an orbital shaker at 200 rpm until the optical density at 600 nm (OD600) reached 0.5. Then the cells were grown an additional 4 h in the presence of varying concentrations of ZnCl2 to evaluate the dynamics of the ZraSR TCS. These cells were used for further studies. After 4 h, cells were harvested by centrifugation for total RNA preparation using the RNeasy Mini kit according to the manufacturer’s instructions (Qiagen) followed by DNase treatment. Reverse transcription was performed with a cDNA synthesis kit (Applied Biosystems) using a random hexamer primers mix according to the manufacturer’s instructions. Specific primers were designed with OLIGO software (version 5.0; Molecular Biology Insights, Cascade, CO, USA) for quantitative expression of the zraP gene and 16sRNA (Table 2). Samples for which the RT step was omitted were used as negative controls to check that the extracted RNA was not contaminated with DNA. Real-time quantitative PCR reactions were performed on the Mini opticon detection system using the SYBR Green PCR Master mix as recommended by the manufacturer. Each quantitative RT-PCR experiment was performed in triplicate for biological samples using separate cultures grown under identical conditions (n = 3) and were calculated automatically by the Mini-opticon software using 16sRNA as an internal control [21].
The expression of the zraP gene was also measured by GFP fluorescence. Cell growth was monitored by the measurement of optical density at 600 nm with a spectrophotometer (Shimadzu, Japan). The fluorescence of GFP-producing cells that were grown in culture was measured using a RF-5301PC spectrofluorimeter (Shimadzu, Japan). The excitation wavelength of the spectrofluorimeter was set at 490/10 nm, and the emission wavelength was set at 510/10 nm. E. coli carrying pZGFP1 without the promoter/operator of the zraP gene was used as a baseline reference to zero the instrument. Raw fluorescence values were expressed in the instrument’s arbitrary relative fluorescence units. The specific fluorescence intensity (SFI) is defined as the raw fluorescence intensity expressed in relative fluorescence units divided by the optical density at 600 nm measured at each time point. At least three measurements were obtained for each sample.
To take photos, cells were screened for fluorescence with a 100× objective on a reflected fluorescence microscope (Olympus, Japan) with a cooled charge-coupled device camera (B&W SenSys, KAF1401). Emission intensity was recorded using MetaMorph image analysis software (Molecular devices, Silicon Valley, CA, USA) with excitation and emission filter sets optimized for EGFP imaging.
Evaluation of metal bioadsorption by engineered bacteria
E. coli strains harboring the pZZ1056 plasmid were grown separately overnight at 37 °C. The overnight culture was diluted 100-fold in fresh LB medium supplemented with 100 μg/ml amplicillin and was incubated at 37 °C in an orbital shaker at 250 rpm. When the cell concentration (OD600) reached 0.5, the cells were induced with varying amounts of 0.5 and 1.0 mM ZnCl2 for 4 h. Then the cells were washed twice with 0.85% NaCl and were resuspended in 0.85% (w/v) NaCl (pH 5.8) to give a cell concentration of 10 g DCW/l. The concentrated cells were incubated in 100 μM ZnCl2 solution for 2 h to evaluate zinc absorption ability and were washed again with 0.85% NaCl. They were then treated with 5 mM EDTA on ice for 30 min to remove the cell surface-bound Zn2+, and the resulting concentration of Zn2+ in the supernatant was measured directly by inductively Coupled Plasma-Mass Spectrometry (ICP, HP4500, Yokogawa, Tokyo, Japan).
Results
Expression monitoring of zraP gene
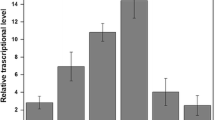
Quantitative RT-PCR was carried out to determine whether the response of Zn2+ concentration on engineered bacteria activated expression of the zraP gene by the ZraRS TCS. RNA samples from E. coli cultures grown for 4 h in nutrient rich and minimal media supplemented with varying concentration of Zn2+ were used, and the results obtained are shown in Fig. 3. Our findings suggest that the transcript level of the zraP gene significantly increased in cells grown at elevated Zn2+ concentrations in both the rich and minimal media. As the zinc ion concentration increased in both cultured media, a similar pattern of zraP gene expression was observed. At 0.1 mM induction, the amount of increase in the zraP transcriptional level was not significant (P < 0.05, n = 3) compared to control, but for zinc ion induction at 1 mM, approximately 8- and 10-fold significant increases (P < 0.05, n = 3) were seen in LB and M9 media, respectively. Thus, these results indicate that the transcriptional level of the zraP gene was induced in the presence of zinc ion at higher concentrations compared with control.
Comparative study of transcriptional levels of the zraP gene in LB (black) and M9 (white) medium after a 4 h Zn2+ exposure. After exposure, the C t value was normalized using the 16sRNA C t value as an internal control. The error bars indicate one standard deviation from the mean. The data are aggregate results from replicated experiments (n = 3)
A zraP dependent reporter plasmid that activates GFP in response to exogenous zinc was constructed. E. coli cells containing the reporter plasmid pZGFP1 exhibited fluorescence in response to the presence of ZnCl2 added to the prescribed medium (Fig. 2b). The stability of the fluorescence signal was estimated by measuring the fluorescence at different time intervals as shown in Fig. 4 for both nutrient rich media and minimal media. The intensity of fluorescence with respect to concentration of zinc in the media was also estimated by the addition of ZnCl2. Fluorescence was observed to be a maximum after 2 h of induction in the nutrient rich culture medium for all zinc ion concentrations. The fluorescence property was found to increase with time. In contrast, maximum fluorescence was exhibited by the bacteria in minimal medium at 0.5 and 1 mM for the 4 h induction period. For zinc concentrations less than 0.5 mM, the organisms exhibited maximum fluorescence after a 5 h induction period. This variation was presumably due to components present in the nutrient rich medium containing sufficient metal ion for induction. Thus, the induction time was chosen as 4 h for subsequent experiments.
Time course of GFP fluorescence for an E. coli strain harboring pZGFP1 after induction with varying concentrations of Zn2+ in both a LB and b M9 medium: 0 mM (closed squares); 0.001 mM (closed circles); 0.01 mM (open triangles); 0.1 mM (open circles); 0.5 mM (open squares); 1.0 mM (closed triangles). The data are aggregate results from replicated zinc treatment experiments (n = 3)
A detailed analysis of the responses of the pZGFP1 harboring strain to zinc in both nutrient rich and minimal medium is described in Fig. 5. Analysis of the response of bacteria present in both media for concentrations of zinc above 10−3 mM revealed two interesting observations. First, for maximal fluorescence induction at 2 h, twofold higher fluorescence was observed in the nutrient rich medium than in the minimal medium. Second, the nutrient rich medium showed that the zinc responding strain started detecting zinc at the 100 μM concentration level, whereas the same strain in minimal media was able to detect zinc even at concentrations as low as 1 μM. The fluorescence exhibited by the strains in both media was found to increase up to a 1 mM concentration of zinc beyond which there was no significant change in the observed fluorescence. The correlation between zinc concentrations and relative fluorescence was found to be linear for both media. In particular, for the concentration range between 100–1,000 μM in nutrient rich medium and 10–1,000 μM in minimal medium, the correlation coefficient was estimated to be 0.919 and 0.983, respectively (Fig. 5). Therefore, the more sensitive and more pronounced fluorescence response at elevated zinc concentrations in both media is believed to be due to the higher extracellular levels of zinc detected by the engineered bacterial system. As the concentration of zinc in the medium was increased above 1,000 μM, further increases in the fluorescence was not linear, which may be due to the toxicity of zinc on the bacterial cells (data not shown). Considering the linear correlation and high sensitivity in minimal medium, the zraP promoter based sensing system could be applied as a zinc biosensor.
E. coli strain pZGFP1 GFP fluorescence after 4 h of Zn2+ exposure in both a LB and b M9 medium. The error bars indicate one standard deviation from the mean. Linear response between fluorescence emission of cells carrying pZGFP1 and varying concentrations of zinc ion. The data are aggregate results from replicated zinc treatment experiments (n = 3)
Zinc bioadsorption by engineered bacteria
The results presented above demonstrate that the genetically engineered bacterial cells show a dynamic response on exposure to an exogenous Zn2+ concentration. Based on these results, we next constructed a cell surface bacterial system that helps to selectively adsorb exogenous Zn2+. The ompC coding region was integrated with ZBP, and inserted into the pUHup1 plasmid to construct pZZ1056 in which chimeric protein was under the control of a zraP promoter. In order to evaluate metal bioadsorption by this engineered strain, the strain was grown in LB medium in the presence of 0.5 and 1.0 mM ZnCl2 induction concentrations. As shown in Fig. 6, cells displaying ompC t –ZBP adsorbed Zn2+ with a higher efficiency than cells harboring pUC19. Cells harboring pZZ1056 could adsorb 97.5 and 163.6 μmol of Zn2+ per gram DCW at 0.5 and 1 mM ZnCl2 induction, respectively, while uninduced cell strains harboring pZZ1056 adsorbed 37.5 μmol of Zn2+ per gram DCW. The observed slight increase in Zn2+ adsorption by uninduced cells may be due to a leaking expression of ompC t –ZBP [22]. This enhancement of adsorption ability by induced cells indicated that the displayed ZBP at the cell surface was very stable in the zinc ion trapping process. Regarding the adsorption of other metals, namely Cu2+, by induced cells, the amount recovered from the cell surface was not as great as that of Zn2+ due to the selective metal adsorption peptides on the cell surface.
Discussion
Currently, a number of heavy metal bacterial biosensors have been developed to quantify gene transcript levels on exposure of bacteria to heavy metals [23–26]. Studies have also revealed that selected genes were specifically up-regulated upon exposure of the bacteria to metal ions [27]. In this study, a TCS based zinc monitoring system was developed for assessing exogenous levels of zinc. The engineered zinc sensing system was able to sense zinc ion concentrations from 0.001 mM and higher in minimal media. Thus, toxic levels of zinc in the environment as prescribed by guidelines of the Environmental Protection Agency (5.0 mg/l or 0.030 mM of zinc) could be quantitatively determined by this simple engineered bacterial system, and this could be considered a breakthrough in the field of biosensors for monitoring toxic zinc contamination in exogenous liquid media (aquatic environment).
The amount of Zn2+ adsorbed by various cell surface display systems in E. coli were compared, and the engineered bacterial system accumulated substantially higher amounts of zinc than other bacterial surface display systems [20, 28] (data not shown). The development of the pZGFP1 reporter in conjunction with bioremediation efforts would complement analytical zinc detection methodologies by fast monitoring and removal of toxic levels of bioavailable zinc contamination in an industrial setting. This study found cell surface engineered bacteria were able to absorb extracellular zinc efficiently without additional induction system. These synthetic heavy metal removal bacteria are not only an example of the application of synthetic biology on bioengineering, but also represent a general strategy for developing multifunctional synthetic bacteria systems.
References
Berg JM, Shi Y (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081–1085
Andreini C, Banci L, Bertini I, Rosato A (2006) Zinc through the three domains of life. J Proteome Res 5:3173–3178
Fraústo da Silva JJR WRJP (2001) The biological chemistry of the elements: the inorganic chemistry of life. Oxford University Press, Oxford
Outten CE, O’Halloran VT (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492
Blencowe DK, Morby AP (2003) Zn(II) metabolism in prokaryotes. FEMS Microbiol Rev 27:291–311
Outten CE, Outten FW, O’Halloran TV (1999) DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem 274:37517–37524
Franke S, Grass G, Nies DH (2001) The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965–972
Leonhartsberger S, Huber A, Lottspeich F, Bock A (2001) The hydH/G Genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J Mol Biol 307:93–105
Noll M, Petrukhin K, Lutsenko S (1998) Identification of a novel transcription regulator from Proteus mirabilis, PMTR, revealed a possible role of YJAI protein in balancing zinc in Escherichia coli. J Biol Chem 273:21393–21401
Reitzer L, Schneider BL (2001) Metabolic context and possible physiological themes of {sigma}54-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65:422–444
Beard SJ, Hashim R, Membrillo-Hernandez J, Hughes MN, Poole RK (1997) Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol 25:883–891
Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP (1999) ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol 31:893–902
Rensing C, Mitra B, Rosen BP (1997) The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA 94:14326–14331
Binet MRB, Poole RK (2000) Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase ZntA in Escherichia coli. FEBS Lett 473:67–70
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Miller WG, Leveau JH, Lindow SE (2000) Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250
Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462
Matsubara T, Hiura Y, Kawahito O, Yasuzawa M, Kawashiro K (2003) Selection of novel structural zinc sites from a random peptide library. FEBS Lett 555:317–321
Jeanteur D, Lakey JH, Pattus F (1991) The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol 5:2153–2164
Xu Z, Lee SY (1999) Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Appl Environ Microbiol 65:5142–5147
Eleaume H, Jabbouri S (2004) Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Methods 59:363–370
Kotrba P, Doleckova L, de Lorenzo V, Ruml T (1999) Enhanced bioaccumulation of heavy metal ions by bacterial cells due to surface display of short metal binding peptides. Appl Environ Microbiol 65:1092–1098
Huckle JW, Morby AP, Turner JS, Robinson NJ (1993) Isolation of a prokaryotic metallothionein locus and analysis of transcriptional control by trace metal ions. Mol Microbiol 7:177–187
Ivask A, Virta M, Kahru A (2002) Construction and use of specific luminescent recombinant bacterial sensors for the assessment of bioavailable fraction of cadmium, zinc, mercury and chromium in the soil. Soil Biol Biochem 34:1439–1447
Riether K, Dollard MA, Billard P (2001) Assessment of heavy metal bioavailability using Escherichia coli zntAp:lux and copAp:lux-based biosensors. Appl Microbiol Biotechnol 57:712–716
Tauriainen S, Karp M, Chang W, Virta M (1998) Luminescent bacterial sensor for cadmium and lead. Biosen Bioelectron 13:931–938
Magrisso S, Erel Y, Belkin S (2008) Microbial reporters of metal bioavailability. Microb Biotechnol 1:320–330
Sousa C, Cebolla A, de Lorenzo V (1996) Enhanced metalloadsorption of bacterial cells displaying poly-His peptides. Nat Biotechnol 14:1017–1020
Acknowledgments
This work was supported by the BioGreen 21 Project (PJ008057) of the Rural Development Administration of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ravikumar, S., Yoo, Ik., Lee, S.Y. et al. A study on the dynamics of the zraP gene expression profile and its application to the construction of zinc adsorption bacteria. Bioprocess Biosyst Eng 34, 1119–1126 (2011). https://doi.org/10.1007/s00449-011-0562-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-011-0562-7