Abstract
Environmental gradients play a key role in shaping diversity in tropical forests. However, we have a little understanding of how evolutionary diversity is affected by gradients and the role of niche persistence in flooded forests in dry biomes. Here, we assessed the evolutionary diversity across a flooding gradient in the Caatinga Domain of South America. We established 120 plots across four tributaries of the São Francisco River, eastern Brazil, consisting of 72 plots in flooded, 24 in occasionally flooded, and 24 in unflooded forests. We computed richness, phylogenetic diversity (PD), mean nearest taxon distance (MNTD), and mean pairwise phylogenetic distance (MPD) and their standardized effect sizes (ses.PD, ses.MNTD, and ses.MPD). We found low richness, low PD, and high MNTD values in flooded forests relative to unflooded and occasionally flooded forests. MPD did not differ across the flooding gradient. The standardized effect size metrics were higher in flooded forests. Despite the unflooded and occasionally flooded forests being rich in terms of species and correlated phylogenetic structure, flooded forests showed more lineage diversity than expected by chance. We assessed whether this pattern could be driven by resprouting ability testing its phylogenetic signal. Resprouting is randomly distributed across phylogeny, but plant communities are likely assembled from random draws of the resprouters’ lineage pool. Quantifying evolutionary diversity across flooding gradients in dry environments brought new insights to how the same environmental filters may lead to disparate patterns of evolutionary diversity and the role of response traits in allowing certain clades to persist in flooded habitats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental gradients act as filters on the distribution of biodiversity across multiple spatial scales (Carvajal-Endara et al. 2017). Gradients associated with the frequency and intensity of flooding are known to shape the structure and dynamics of seasonally flooded plant communities, as well as their floristic composition, species richness, and functional diversity across wide geographical ranges (Capon 2005; Budke et al. 2007, 2008, 2010; Gonzalez et al. 2010; Wittmann et al. 2010; Kotowski et al. 2010; Violle et al. 2010; Wittmann et al. 2013; Moreira et al. 2015; Moor et al. 2015; Luo et al. 2016). Increased flooding should decrease taxonomic diversity (Budke et al. 2008) by selecting for a subset of species that possess the traits necessary to survive under higher flooding stress (Giehl and Jarenkow 2015). Environmental filters can also affect evolutionary diversity. For example, the influence of stress gradients on overall patterns of evolutionary diversity has been documented in seasonally dry tropical forests (drought stress; Pennington et al. 2009), savannas in Central Brazil, and white-sand forests in the Amazon (edaphic-related stress, Guevara et al. 2016; Meira-Neto et al. 2017), zooplankton metacommunities (anthropogenic disturbances; Gianuca et al. 2017), sponges (light availability gradients; Quattrini et al. 2017), and in seasonally flooded forests (water saturation stress; Giehl and Jarenkow 2015; Aldana et al. 2016).
In South America, studies of evolutionary diversity across flooding gradients have been carried out comparing distinct habitats. For example, Aldana et al. (2016) found more closely related eudicot lineages in seasonally flooded areas relative to unflooded (terra firme) forests in the Amazon, supporting the idea of flooding as a strong environmental filter. In a broad-scale study of environmental gradients, Gonzalez-Caro et al. (2014) found phylogenetic clustering both in dry and floodplain forests in Colombia. These stressful habitats seem to play a substantial role filtering out lineages not adapted to water-related stress. While this phylogenetic clustering in response to hydric stresses is likely to reflect phylogenetic signal in key traits (Webb et al. 2002), it is not known which traits drive this clustering (but see Parolin 2008). However, Giehl and Jarenkow (2015) linked higher phylogenetic richness under the higher levels of water saturation in a subtropical moist forest to patterns in the phylogenetic signal (convergence) of traits (e.g., resprouting ability) that are associated with stress flooding tolerance.
Traits that mediate the response of a species to environment changes (Violle et al. 2007) have helped to understand the link between environmental filters and the response of diversity to environmental stress. For example, fire can filter species according to their bark traits (Charles-Dominique et al. 2017), with thicker barked species more likely to survive fire. For physical disturbances such as floods, traits that allow an individual to recover from physical damage and thus persist in situ (their persistence niche) will be important. Resprouting ability is likely to be a key trait in determining persistence, as it determines the ability of a tree to survive and recover following trunk breakage (Bond and Midgley 2001; Araujo et al. 2017). Evidence that resprouting ability confers tolerance to flooding events comes from observations that multi-stemmed trees are favored under regular disturbance-stress regimes (Bond and Midgley 2003; Dunphy et al. 2000; Bellingham and Sparrow 2009). However, the phylogenetic signal—measure of trait distribution across phylogenetic tree to a given community, or persistence niche strategies, such as resprouting—is poorly known, so the consequences of environmental filtering due to flooding stress for the phylogenetic structure of tree communities are unclear.
No study to date has assessed the importance of flooding as an environmental filter in seasonally dry biomes—flooded forests subjected to < 1800 mm rainfall per year, the typical annual rainfall of the tropical dry forests (Dryflor 2016; but see Gonzalez-Caro et al. 2014 for phylogenetic diversity comparisons across biomes). Here, we aim to assess the influence of flooding gradients on evolutionary diversity across the South American Caatinga Domain, the largest continuous extent of the tropical dry forest biome (Fernandes 2003; Santos et al. 2012). While dry forests and woodlands, the predominant vegetation type in the Caatinga Domain and in tropical dry forest as a whole, have natural constraints related to water shortage, seasonally flooded forests in tropical dry forest are also potentially affected by soil waterlogging. Thus, we have a natural scenario for testing the idea that stress gradients act as environmental filters to shape patterns of evolutionary diversity, and to examine the role of persistence strategy in influencing such patterns. Here, we hypothesize that water-related stresses impose a strong environmental filter, thus leading to the similar patterns of evolutionary diversity. We expect that both hydric stresses, waterlogging and water shortage, will lead to low evolutionary diversity, because selected lineages could persist in such conditions in a dry climate. In addition, as multi-stemmed trees are favored in harsh conditions, phylogenetic signal of resprouting ability could explain such evolutionary patterns, in flooded and dry forests. Finally, we predicted that intermittently flooded forests would show similar evolutionary diversity patterns to dry forests, because we expect that drought stress imposes the dominant environmental filter on the lineage pool, with environmental filters imposed by flooding further modifying the lineages present.
Methods
Study area and sampling
Sampling units in this study were tree communities across flooding gradients in four tributaries of the São Francisco River basin, eastern Brazil (Fig. 1). In each area, we established 30 plots of 20 × 20 m (400 m2), totaling 120 plots and 4.8 ha. In some cases, plots were 10 × 40 m (400 m2) due to narrower vegetation bands. The plots were then classified into three flooding degrees, according to their distance to the river channel, with 72 plots in the areas that were regularly flooded (hereafter flooded), 24 plots that experienced occasional flooding (occasionally flooded), and 24 plots that did not experience flooding (unflooded). All the tree individuals with diameter at breast height (DBH) ≥ 3.2 cm (≥ 10 cm circumference at breast height; CBH—standard criteria for dry forests) were measured and identified at the species level during sampling. Our classification of flooding degree was based on local knowledge. For instance, our flooded forests, located by riparian habitats, are annually flooded, have evidence of sedimentation deposits, and also short-lived ponds. The occasionally flooded forests are characterized by shallow depressions and are flooded at a frequency of about once in every 30 years. Finally, the unflooded forests are characterized by typical dry forests, with no evidence of temporary ponds. All plot censuses were performed during the dry seasons (July–November 2012, 2013, and 2015). The tree species were identified in the field and nomenclature was revised in REFLORA database, accessed at 20 July 2017.

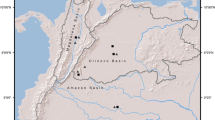
Adapted from Pereira 2013
Map showing the location of the sampling areas (labeled C, V, P, and J) across the São Francisco River basin, Minas Gerais and Bahia states, Brazil. C Carinhanha river, V Verde Grande river, P Paracatu river, J Jequitaí river. Vegetation types found across the flooding gradients; classified according to the flooding frequency (see “Methods”). Dashed lines show water level during the flooding season.
Phylogenetic diversity metrics
A phylogenetic tree of the whole species pool was generated using Phylomatic in Phylocom version 4.2 (Webb et al. 2008). This tool provides a phylogenetic hypothesis for the relationships among taxa by matching the list of species, with up-to-date family and genus names, and tip labels of a provided megatree (Webb and Domoghue 2005). In this case, the topology of R20120829.new provided at http://phylodiversity.net/phylomatic/ was used. An ultrametric phylogeny including branch length in millions of years (Ma) was obtained using the branch length adjustment (bladj) in Phylocom. This function fixes the root node (angiosperms, 179 Ma) and other nodes to specified ages based on Wikstrom et al. (2001). We then used the Phylocom phylogeny, which includes all the species in our data set, to calculate six metrics that evaluate the evolutionary history in communities: (1) the total phylogenetic branch length of all species occurring in a given community, i.e., phylogenetic diversity sensu stricto (PD; Faith 1992); (2) mean pairwise phylogenetic distance based on branch length (MPD; Webb 2000; Webb et al. 2002); (3) mean nearest taxon distance (MNTD; Webb 2000; Webb et al. 2002); and (4, 5, and 6) the standardized effect sizes for species richness (ses.PD, ses.MPD, and ses.MNTD). Both MPD and MNTD are weighted by abundance and measure the average pairwise distance of all the individuals and between an individual and the most closely related excluding conspecifics, respectively.
Because these standardized metrics have an expected value of 0 and a SD of 1, values < − 1.96 or > 1.96 represent communities that show lower or higher phylogenetic diversity than would be expected by chance, respectively (Kembel et al. 2010), while values within this range indicate that phylogenetic diversity is no different than random expectation, i.e., communities are assembled from random draws of the phylogeny (Kembel et al. 2010).
For each community, these standardizations were accomplished by randomly drawing the same number of species from the phylogeny as present in the community, and repeating this 1000 times. Then, we calculated PD, MPD and MNTD for each randomization taking the difference between the observed value of PD, MPD, and MNTD, the mean of the random values, and dividing these differences by the standard deviation across the randomizations.
Because flooded environments have more sampling units, which could potentially bias analyses and interpretation, we applied the same analysis using an equal number of sampling units per flooding category, and found the same results (Online Resource 1). We computed the phylogenetic diversity analyses using the picante package (Kembel et al. 2010) in the R Statistical Environment.
Data analysis
We tested whether different flooding frequencies lead to significantly distinct amounts of phylogenetic diversity by applying Kruskal–Wallis tests (due to non-normally distributed residuals), followed by post hoc Dunn tests with Bonferroni correction (Dinno 2017). We tested whether using an unresolved phylogeny affects the overall results for phylogenetic diversity by generating 1000 randomly resolved phylogenies, using the ape package (Paradis et al. 2004) in R. We then calculated ses.PD values using all fully resolved phylogenies and correlated the mean values with the ses.PD values found for the unresolved phylogeny. We also used the fully resolved phylogenies to estimate phylogenetic signal in resprouting ability using Blomberg’s K (Blomberg et al. 2003), which quantifies the amount of variance in an observed trait in relation to the expected variance under a Brownian motion model of evolution. We assessed the significance of phylogenetic signal by recalculating K a thousand times on phylogenies with randomly permuted tips. Finally, we mapped relative abundance in seasonally flooded sites on to the unresolved phylogeny by interpolating states at internal branches through a maximum-likelihood ancestral state reconstruction (Schluter et al. 1997). We then tested whether the relative abundance of lineages found in seasonally flooded sites is associated with resprouting ability using a phylogenetic generalized least-squares approach available in the nlme package (Pinheiro et al. 2017) in R. We conducted the trait mapping and phylogenetic signal analyses using the phytools package (Revell 2012) in R.
Results
Taxonomic and phylogenetic diversity
The three habitat types differed significantly from each other in richness. Unflooded forests had the highest species richness, followed by occasionally flooded and flooded forests (Kruskal–Wallis \(X_{38.77}^{2}\), P < 0.001). The highest phylogenetic diversity (PD), the sum of phylogenetic branch lengths in a community, was found in unflooded and occasionally flooded forests (Kruskal–Wallis \(X_{31.75}^{2}\), P < 0.001) compared to flooded forest, and mirroring the overall patterns of species richness (Online Resource 2, Fig. 2). In contrast, mean nearest taxon distance (MNTD) was significantly higher in flooded forest than occasionally and unflooded forests (Kruskal–Wallis \(X_{25.47}^{2}\), P < 0.001). Finally, mean pairwise distance (MPD) did not differ significantly among the three groups (Fig. 2).
Distribution of species richness (a) and phylogenetic diversity values (PD, MNTD, and MPD, b–d) in tree communities found across flooding gradients of the São Francisco River basin, eastern Brazil. PD phylogenetic diversity, MNTD mean nearest taxon distance, MPD mean pairwise phylogenetic distance. Same letters in two or more groups (flooding frequencies) indicate that means are no different than random expectation (i.e., P > 0.05 in Kruskal–Wallis and post hoc Dunn test with Bonferroni correction)
Lineage diversity
The highest lineage diversity (i.e., ses.PD) was found in flooded and occasionally flooded forests compared to unflooded forests (sesPD; F9279, P < 0.001) (Fig. 3a). These results were robust to phylogenetic uncertainty; the correlation between average ses.PD (from randomly generating 1000 fully bifurcating trees) and ses.PD of the original tree indicates that the lack of phylogenetic resolution in the phylogenetic tree did not affect the overall results (Online Resource 3). Flooded forests had the highest amounts of both neighbor lineage diversity (MNTD standardized for species richness; ses.MNTD), (Kruskal–Wallis \(X_{17.20}^{2}\), P < 0.001, Fig. 3b) and clade lineage diversity (MPD standardized for species richness; ses.MPD, Fig. 3c) compared to occasionally and unflooded forests. Over 7% of the species in flooded forests belong to basal angiosperm lineages (magnoliids), with much lower proportions in occasionally flooded (1.83%) and unflooded forests (2.32%).
Distribution of phylogenetic diversity values standardized for species richness (ses.PD, ses.MNTD, and ses.MPD, a–c) in tree communities found across flooding gradients in the São Francisco River basin, eastern Brazil. ses.PD standardized phylogenetic diversity, ses.MNTD standardized mean nearest taxon distance, ses.MPD standardized mean pairwise phylogenetic distance. Same letters in two or more groups (flooding frequencies) indicate that means are no different than random expectation (i.e., P > 0.05 in Kruskal–Wallis and post hoc Dunn test with Bonferroni correction)
Resprouting ability
The phylogenetic generalized least-squares (PGLS) model revealed higher lineage abundance in seasonally flooded environments for lineages with resprouting ability (Fig. 4; Delta AIC = − 14.617), but resprouting ability has no phylogenetic signal. This result is robust to phylogenetic uncertainty (assessed by calculating phylogenetic signal after randomly generating 1000 fully bifurcating trees, Online Resource 4).
Time-calibrated phylogeny of angiosperm species in tree communities found across flooding gradients of the São Francisco River basin, eastern Brazil. Colours represent the relative abundance of lineages found in seasonally flooded sites (AF); warmer colours indicate higher values. The figure illustrates the phylogenetic overdispersion of lineages that are abundant in seasonally flooded sites. Bars represent a resprouting index and larger bars indicate higher resprouting ability. Scale (length) is in Ma and is equivalent to branch lengths in the phylogeny (50 Ma)
Discussion
We had hypothesized that water-related stress imposes a strong environmental filter, resulting in low evolutionary diversity in both flooded and drought habitats, but differences in diversity patterns between different unstandardized phylogenetic diversity metrics (PD, MNTD, and MPD) were found across the flooding gradient. In addition, higher amounts of lineage diversity (ses.PD, ses.MNTD, and ses.MPD) under higher levels of seasonal flooding indicate that flooded forests show greater lineage diversity than expected given their richness and the establishment of dry forest lineages does not occur randomly across the flooding gradient. Though, in moist biomes, evolutionary diversity is negatively correlated with flooding frequency (Aldana et al. 2016), we found that, in dry biomes, lineage diversity is higher in flooded forests. Finally, we found that the most abundant lineages persist in seasonally flooded environments by resprouting, a response trait to the environment that was randomly distributed in the phylogeny.
Phylogenetic and lineage diversity
Raw phylogenetic diversity shows intriguing patterns across the flooding gradient. First, occasionally flooded and unflooded forests have similar patterns, which confirmed the assumption that both forests are ecologically and evolutionary assembled by drought stress selecting drought tolerant clades. These forests showed high species diversity, high PD, and low MNTD values compared to flooded forests with low PD and high MNTD values. It is possible to explain these patterns due to correlation between metrics (Online Resource 2). Richness was positively correlated with phylogenetic diversity not controlled for species richness (PD), but negatively correlated with MNTD. Increasing richness increases the sum of the branch length of the phylogeny (PD), but decreases mean distance between family and genus-level clades (MNTD). Thus, it is possible that drought tolerant clades in unflooded forests are old and confined to tropical dry forest clades (Pennington et al. 2009). It seems that unflooded forest communities have only evolved a few times (low lineage diversity: low ses.PD, ses. MNTD, and ses.MPD), but have recent and great diversification (high richness and PD values). This is consistent with the previous assessments of the non-flooded landscapes within the dry forest biome (Pennington et al. 2009; Gonzalez-Caro et al. 2014). In contrast, flooding seems to filter many distantly related flood-tolerant lineages (high lineage diversity: high ses.PD, ses.MNTD, and ses.MPD) related to resprouting ability, but each lineage has low species richness and correlated PD values, with ancient diversification (high MNTD and MPD values). These clades could represent a flood-tolerant subset of the vast and widespread pool of mesic-adapted taxa distributed throughout tropical rainforests, including the Amazon Basin.
Two factors emerge to explain this pattern. First, gallery forests in dry biomes seem to connect tropical moist biomes (e.g., Amazon Basin and Atlantic forest sensu stricto) as relictual migratory routes (Oliveira-Filho and Ratter 1995; Batalha-Filho et al. 2013). Second, tropical rainforests are considered older moist biomes compared to dry forests, and, therefore, have more lineages (Eiserhardt et al. 2017). If it is correct, flooded forests could be seen as a refuge for mesic-adapted taxa in dry biomes, particularly tropical dry forest.
On one hand, there is compelling evidence in the literature supporting Phylogenetic Niche Conservatism for warm environments; only a small subset of lineages was able to colonize frost-affected environments over evolutionary time (Latham and Ricklefs 1994; Jablonski et al. 2006; Wiens and Donoghue 2004; Wiens et al. 2010; Zanne et al. 2014; Kerkhoff et al. 2014; Augusto et al. 2014). On the other hand, patterns of Phylogenetic Niche Conservatism for mesic, moist environments are still unclear and poorly explored (Qian et al. 2017; Qian and Sandel 2017), and our results bring further support to the idea that seasonally dry environments are mainly assembled from a subset of plant lineages that can tolerate drought. While tree communities in seasonally flooded forests are found under the same precipitation conditions as unflooded, dry forests (6 months with less than 100 mm of total rainfall), a higher level of soil moisture during the dry season, due to proximity to the river channel, seems to provide a suitable environment for most plant lineages in the phylogeny, thus leading to the observed patterns of higher lineage diversity. Moreover, high lineage diversity (ses.PD) was found in occasionally flooded forest, which suggests that even infrequent flooding could allow mesic-adapted taxa to establish in drought environment. This phylogenetic impoverishment in unflooded dry forests is even more evident in basal lineages (magnoliids), which are known to conserve mesic habitat preferences (Coronado et al. 2015; Carlucci et al. 2017).
Resprouting ability
We found that resprouting ability influences association with flooded forests, but, as it is randomly distributed across the phylogeny, environmental filtering due to resprouting ability does not lead to a loss of evolutionary diversity. This contrasts the phylogenetic overdispersion (convergence) for resprouting ability in tree communities found across flooding gradients in the Atlantic Forest Domain (Giehl and Jarenkow 2015), and suggests that the mechanisms driving the patterns of community assembly are overall distinct in seasonally dry environments. In our study, because flooding is filtering for plant lineages that have the ability to resprout after flooding disturbance, and resprouting is not a phylogenetically conserved trait, plant communities in seasonally flooded environments are likely assembled from random draws of the resprouting lineage pool. This leads to the patterns of evolutionary diversity that are not different than random expectation (see Fig. 2 and “Methods”), but overall higher than in unflooded, dry forests, which are assembled from an impoverished lineage pool that can tolerate harshness associated with water shortage.
Conclusions
Quantifying evolutionary diversity across flooding gradients in seasonally dry environments brought new insights on how the same environmental filters may lead to disparate patterns of evolutionary diversity. Dry forests are composed of a subset of clades that can tolerate drought, with high species diversity and correlated phylogenetic diversity in some specific confined clades, with intermittently flooded forests showing similar patterns. Besides high lineage diversity, flooded forests are composed of distantly related flood-tolerant lineages with less species diversity, but high family and genus-level phylogenetic diversity. Although resprouting is a randomly distributed trait across the phylogeny, a wide range of lineages that have the ability to resprout can persist in flooded habitats. Thus, flooded forests are dominated by resprouters. Multi-stemmed trees in the persistence niche are selected for in a lineage pool that can resprout in an unstable environment.
References
Aldana AM, Carlucci MB, Fine PVA, Stevenson PR (2016) Environmental filtering of eudicot lineages underlies phylogenetic clustering in tropical South American flooded forests. Oecologia 183(2):327–335. https://doi.org/10.1007/s00442-016-3734-y
Araujo FDC, Mendes CN, Pires GG, Menino GCO, Fontes MAL, Santos RM (2017) Adaptive strategy of tree communities on an environmental harshness hinterland inselberg in Minas Gerais, Brazil. Aust J Bot. https://doi.org/10.1071/BT16252
Augusto L, Davies TJ, Delzon S, Schrijver ADE (2014) The enigma of the rise of angiosperms: can we untie the knot? Ecol Lett 17:1326–1338
Batalha-Filho H, Fjeldsa J, Fabre P-H, Myiaki CY (2013) Connections between the Atlantic and the Amazonian forest avifaunas represent distinct historical events. J Ornithol 154:41–50
Bellingham PJ, Sparrow AD (2009) Multi stemmed trees in montane rain forests: their frequency and demography in relation to elevation, soil nutrients and disturbance. J Ecol 97:472–483. https://doi.org/10.1111/j.1365-2745.2009.01479.x
Blomberg SP, Garland T Jr, Ives AR (2003) Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57(4):717–745
Bond WJ, Midgley JJ (2001) Ecology of sprouting in woody plants: the persistence niche. Trends Ecol Evol 16:45–51. https://doi.org/10.1016/S0169-5347(00)02033-4
Bond WJ, Midgley JJ (2003) The evolutionary ecology of sprouting in woody plants. Int J Plant Sci 164:S103–S114. https://doi.org/10.1086/374191
Budke JC, Jarenkow JA, Oliveira-Filho AT (2007) Relationships between tree component structure, topography and soils of a riverside forest, Rio Botucaraí, Southern Brazil. Plant Ecol 189:187–200. https://doi.org/10.1007/s11258-006-9174-8
Budke JC, Jarenkow JA, Oliveira-Filho AT (2008) Tree community features of two stands of riverine forest under diferente flooding regimes in Southern Brazil. Flora 203:162–174
Budke JC, Jarenkow JA, Oliveira-Filho AT (2010) Intermediary disturbance increases tree diversity in riverine forest of southern Brazil. Biodivers Conserv 19:2371–2387. https://doi.org/10.1007/s10531-010-9845-6
Capon SJ (2005) Flood variability and spatial variation in plant community composition and structure on a large arid floodplain. J Arid Environ 60:283–302. https://doi.org/10.1016/j.jaridenv.2004.04.004
Carlucci MB, Seger GDS, Sheil D, Amaral IL, Chuyong GB, Ferreira LV, Galatti U, Hurtado J, Kenfack D, Leal DC, Lewis SL, Lovett JC, Marshall AR, Martin E, Mugerwa B, Munishi P, Oliveira ÁCA, Razafimahaimodison JC, Rovero F, Sainge MN, Thomas D, Pillar VD, Duarte LDS (2017) Phylogenetic composition and structure of tree communities shed light on historical processes influencing tropical rainforest diversity. Ecography 40:521–530. https://doi.org/10.1111/ecog.02104
Carvajal-Endara S, Hendry AP, Emery NC, Davies TJ (2017) Habitat filtering not dispersal limitation shapes oceanic island floras: species assembly of the Galápagos archipelago. Ecol Lett 20(4):495–504. https://doi.org/10.1111/ele.12753
Charles-Dominique T, Midgley JF, Bond WJ (2017) Fire frequency filters species by bark traits in a savanna-forest mosaic. J Veg Sci 28:728–735
Coronado HEN, Dexter KG, Pennington RT, Chave J, Lewis SL, Alexiades MN, Alvarez E, Alves de Oliveira A, Amaral IL, Araujo-Murakami A, Arets EJMM, Aymard GA, Baraloto C, Bonal D, Brienen R, Cerón C, Cornejo Valverde F, Di Fiore A, Farfan-Rios W, Feldpausch TR, Higuchi N, Huamantupa-Chuquimaco I, Laurance SG, Laurance WF, López-Gonzalez G, Marimon BS, Marimon-Junior BH, Monteagudo Mendoza A, Neill D, Palacios Cuenca W, Peñuela Mora MC, Pitman NCA, Prieto A, Quesada CA, Ramirez Angulo H, Rudas A, Ruschel AR, Salinas Revilla N, Salomão RP, Segalin de Andrade A, Silman MR, Spironello W, ter Steege H, Terborgh J, Toledo M, Valenzuela Gamarra L, Vieira ICG, Vilanova Torre E, Vos V, Phillips OL (2015) Phylogenetic diversity of Amazonian tree communities. Divers Distrib 21:1295–1307
Dinno A (2017) Dunn.test: Dunn’s test of multiple comparisons using rank sums. R package version 1.3.3. https://CRAN.R-project.org/package=dunn.test. Accessed 7 Jan 2017
Dryflor (2016) Plant diversity patterns in neotropical dry forests and their conservation implications. Science 353:1383–1387
Dunphy BK, Murphy PG, Lugo AE (2000) The tendency for trees to be multiple-stemmed in tropical and subtropical dry forests: studies of Guanica Forest, Puerto Rico. Trop Ecol 41:161–167
Eiserhardt WL, Couvreur TLP, Baker WJ (2017) Plant phylogeny as a window on the evolution of hyperdiversity in the tropical rainforest biome. New Phytol. https://doi.org/10.1111/nph.14516
Faith DP (1992) Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10
Fernandes A (2003) Conexões florísticas do Brasil. Banco do Nordeste, Fortaleza, Brazil, 1st edn. Banco do Nordeste, Fortaleza
Gianuca AT, Engelen J, Brans KI, Hanashiro FTT, Vanhamel M, van den Berg EW, Souffreau C, De Meester Luc (2017) Taxonomic, functional and phylogenetic metacommunity ecology of cladoceran zooplankton along urbanization gradientes. Ecography 40:001–011. https://doi.org/10.1111/ecog.02926
Giehl EL, Jarenkow JA (2015) Disturbance and stress gradients result in distinct taxonomic, functional and phylogenetic diversity patterns in a subtropical riparian tree community. J Veg Sci 26(5):889–901. https://doi.org/10.1111/jvs.12288
Gonzalez MA, Roger A, Courtois EA, Jabot F, Norden N, Paine CET, Baraloto C, Thébaud C, Chave J (2010) Shifts in species and phylogenetic diversity between sapling and tree communities indicate negative density dependence in a lowland rain forest. J Ecol 98:137–146
Gonzalez-Caro S, Umaña MN, Stevenson PR, Swenson NG (2014) Phylogenetic alpha and beta diversity in tropical tree assemblages along regionalscale environmental gradients in northwest south America. J Plant Ecol 7:145–153
Guevara JE, Damasco G, Baraloto C, Fine PVA, Peñuela MC, Castilho C, Vincentini A, Cárdenas D, Wittmann F, Targhetta N, Phillips O, Stropp J, Amaral I, Maas P, Monteagudo A, Jimenez EM, Thomas R, Brienen R, Duque A, Magnusson W, Ferreira C, Honorio E, Matos FA, Arevalo FR, Engel J, Petronelli P, Vasquez R, ter Steege H (2016) Low phylogenetic beta diversity and geographic neo-endemism in Amazonian white-sand forests. Biotropica 48(1):34–46. https://doi.org/10.1111/btp.12298
Jablonski D, Roy K, Valentine JW (2006) Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science 314:102–106
Kembel SW, Cowan PD, Helmus MR et al (2010) Picante: r tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. https://doi.org/10.1093/bioinformatics/btq166
Kerkhoff AJ, Moriarty PE, Weiser MD (2014) The latitudinal species richness gradient in New World woody angiosperms is consistent with the tropical conservatism hypothesis. Proc Natl Acad Sci USA 111:8525–8530
Kotowski W, Beauchard O, Opdekamp W, Meire P, van Diggelen R (2010) Waterlogging and canopy interact to control species recruitment in floodplains. Funct Ecol 24:918–926. https://doi.org/10.1111/j.1365-2435.2009.01682
Latham RE, Ricklefs RE (1994) Continental comparisons of temperate-zone tree species diversity. In: Ricklefs RE, Schluter D (eds) Species diversity in ecological communities. University of Chicago Press, Chicago, pp 294–314
Luo FL, Huang L, Lei T, Xue W, Li HL, Yu FH, Cornelissen JHC (2016) Responsiveness of performance and morphological traits to experimental submergence predicts field distribution pattern ofwetland plants. J Veg Sci 27:340–351. https://doi.org/10.1111/jvs.12352
Meira-Neto JAA, Tolentino GS, Silva MCNA, Neri AV, Gastauer M, Magnago LFS, Yuste JC, Valladares F (2017) Functional antagonism between nitrogen-fi xing leguminous trees and calcicole-drought-tolerant trees in the Cerrado. Acta Bot Bras 31(1):11–18. https://doi.org/10.1590/0102-33062016abb0380
Moor H, Hylander K, Norberg J (2015) Predicting climate change effects on wetland ecosystem services using species distribution modeling and plant functional traits. Ambio 44(1):113–126. https://doi.org/10.1007/s13280-014-0593-9
Moreira SN, Eisenlohr PV, Pott A, Pott V, Oliveira-Filho AT (2015) Similar vegetation structure in protected and non-protected wetlands in Central Brazil: conservation significance. Environ Conserv 42(4):356–362. https://doi.org/10.1017/S0376892915000107
Oliveira-Filho AT, Ratter JA (1995) A study of the origin of Central Brazilian forests by the analysis of plant species distribution patterns. Edinb J Bot 52(2):141–194
Paradis E, Claude J, Strimmer K (2004) APE: analisys of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290
Parolin P (2008) Submerged in darkness: adaptations to prolonged submergence by woody species of the Amazonian floodplains. Ann Bot 103:359–376. https://doi.org/10.1093/aob/mcn216
Pennington RT, Lavin M, Oliveira-Filho AT (2009) Woody plant diversity, evolution, and ecology in the tropics: perspectives from seasonally dry tropical forests. Ann Rev 40:437–457. https://doi.org/10.1146/annurev.ecolsys.110308.120327
Pereira DGS (2013) Conexões estruturais e florísticas de ecounidades da vegetação arbórea de uma planície inundável do Médio São Francisco. PhD dissertation, Department of Forestry, Federal University of Lavras, Lavras, Minas Gerais, Brazil
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team (2017) nlme: Linear and nonlinear mixed effects models. R package version 3.1-131. https://CRAN.R-project.org/package=nlme. Accessed 8 Oct 2017
Qian H, Sandel B (2017) Phylogenetic structure of regional angiosperm assemblages across latitudinal and climatic gradients in North America. Global Ecol Biogeogr 26:1258–1269
Qian H, Jin Y, Ricklefs RE (2017) Patterns of phylogenetic relatedness of angiosperm woody plants across biomes and life-history stages. J Biogeogr. https://doi.org/10.1111/jbi.12936
Quattrini AM, Gómez CE, Cordes EE (2017) Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 183:221–236. https://doi.org/10.1007/s00442-016-3765-4
Revell LJ (2012) phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3(2):217–223
Santos RM, Oliveira-Filho AT, Eisenlohr PV, Queiroz LP, Cardoso DBOS, Rodal MJN (2012) Identity and relationships of the Arboreal Caatinga among other floristic units of seasonally dry tropical forests (SDTFs) of north-eastern and Central Brazil. Ecol Evol 2(2):409–428. https://doi.org/10.1002/ece3.91
Schluter D, Price T, Mooers AO, Ludwig D (1997) Likelihood ancestor states in adaptive radiation. Evolution 51(6):1699–1711
Violle C, Navas M-L, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E (2007) Let the concept of trait be functional! Oikos 116:882–892
Violle C, Bonis A, Plantegenest M, Cudennec C, Damgaard C, Marion B, Le Coeur D, Bouzillé JB (2010) Plant functional traits capture species richness variations along a flooding gradient. Oikos 00:001–017. https://doi.org/10.1111/j.1600-0706.2010.18525.x
Webb CO (2000) Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat 152(2):145–155
Webb CO, Domoghue MJ (2005) Phylomatic: tree assembly for applied phylogenetics. Mol Ecol Notes 5(1):181–183
Webb CO, Ackerly DD, McPeek MA, Donoghue MJ (2002) Phylogenies and community ecology. Ann Rev 33:475–505. https://doi.org/10.1146/annurev.ecolsys.33.010802.150448
Webb CO, Ackerly DD, Kembel SW (2008) Phylocom: software for the analysis of phylogenetic community structure and character evolution. Bioinformatics 24(18):2098–2100. https://doi.org/10.1093/bioinformatics/btn358
Wiens JJ, Donoghue MJ (2004) Historical biogeography, ecology and species richness. Trends Ecol Evol 19(12):639–644. https://doi.org/10.1016/j.tree.2004.09.011
Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, Damschen EI, Davies J, Grytnes JA, Harrison SP, Hawkins BA, Holt RD, McCain CM, Stephens PR (2010) Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett 13:1310–1324
Wikstrom N, Savolainen V, Chase MW (2001) Evolution of the angiosperms: calibrating the family tree. Proc R Soc Lond B 268:2211–2220
Wittmann F, Schongart J, Junk WJ (2010) Phytogeography, species diversity, community structure and dynamics of Amazonian floodplain forests. In: Junk WJ, Piedade MTF, Wittmann F, Schongart J, Parolin P (eds) Amazonian floodplain forests: ecophysiology, biodiversity and sustainable management. Ecological studies 210. Springer, Dordrecht, pp 61–102
Wittmann F, Householder E, Piedade MTF, Assis RL, Schongart J, Parolin P, Junk WJ (2013) Habitat specifity, endemism and the neotropical distribution of Amazonian white-water floodplain trees. Ecography 36:690–707. https://doi.org/10.1111/j.1600-0587.2012.07723.x
Zanne AE, Tank DC, Cornwell WK, Eastman JM, Smith SA, FitzJohn RG, McGlinn DJ, O’Meara BC, Moles AT, Reich PB, Royer DL, Soltis DE, Stevens PF, Westoby M, Wright IJ, Aarssen L, Bertin RI, Calaminus A, Govaerts R, Hemmings F, Leishman MR, Oleksyn J, Soltis PS, Swenson NG, Warman L, Beaulieu JM (2014) Three keys to the radiation of angiosperms into freezing environments. Nature 506:89–92
Acknowledgements
We thank CAPES (Coordenação de Aperfeiçoamento Pessoal de Nível Superior), FAPEMIG (Fundação de Amparo à Pesquisa de Minas Gerais), and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for financial support. We thank Dr. Danilo Neves, who helped improve an earlier version of this manuscript and helped with statistical analysis. We thank Dr. Martin Sullivan for providing comments on this work and for editing the manuscript to improve the quality of the English. We thank the editors and four reviewers for their valuable comments, which greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
FDCA and RMS conceived and designed the experiments. FDCA and RMS performed the experiments. FDCA analyzed the data. FDCA and RMS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Casey P. terHorst.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Araújo, F.D.C., Santos, R.M. Different degrees of water-related stress affect evolutionary diversity in a seasonally dry biome. Oecologia 189, 795–802 (2019). https://doi.org/10.1007/s00442-019-04358-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-019-04358-4