Abstract
Increasing prevalence of wildlife disease accentuates the need to uncover drivers of epidemics. Predators can directly influence disease prevalence via density-mediated effects (e.g., culling infected hosts leading to reduced disease prevalence). However, trait-mediated indirect effects (TMIEs) of predators can also strongly influence disease—but predicting a priori whether TMIEs should increase or decrease disease prevalence can be challenging, especially since a single predator may elicit responses that have opposing effects on disease prevalence. Here, we pair laboratory experiments with a mechanistic, size-based model of TMIEs in a zooplankton host, fungal parasite, multiple predator system. Kairomones can either increase or decrease body size of the host Daphnia, depending on the predator. These changes in size could influence key traits of fungal disease, since infection risk and spore yield increase with body size. For six host genotypes, we measured five traits that determine an index of disease spread (R 0). Although host size and disease traits did not respond to kairomones produced by the invertebrate predator Chaoborus, cues from fish reduced body size and birth rate of uninfected hosts and spore yield from infected hosts. These results support the size model for fish; the birth and spore yield responses should depress disease spread. However, infection risk did not decrease with fish kairomones, thus contradicting predictions of the size model. Exposure to kairomones increased per spore susceptibility of hosts, countering size-driven decreases in exposure to spores. Consequently, synthesizing among the relevant traits, there was no net effect of fish kairomones on the R 0 metric. This result accentuates the need to integrate the TMIE-based response to predators among all key traits involved in disease spread.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Increases in disease prevalence in wildlife populations have created a great deal of concern among ecologists and managers (Harvell et al. 1999; Daszak et al. 2000; Dobson and Foufopoulos 2001; Kilpatrick 2011). What drives this increase in parasite outbreaks? We now know that the interactions of hosts and/or parasites with other species can strongly influence disease spread (Hatcher et al. 2006; Keesing et al. 2006, 2010). These discoveries provide predictive insight into variation of epidemic size but also into management options to control outbreaks (Ostfeld and Holt 2004; Keesing et al. 2006). For instance, theory and data reveal that predators reduce disease prevalence by culling hosts directly; this matters because spread and prevalence of disease often increase with host density (Packer et al. 2003; Ostfeld and Holt 2004; Hall et al. 2005). Perhaps more importantly, predators often selectively cull infected hosts, further inhibiting epidemics (Packer et al. 2003; Ostfeld and Holt 2004). These two mechanisms suggest that predator management could reduce disease. However, the density of some predators can correlate positively with the size of epidemics—they might instead spread disease (Choisy and Rohani 2006; Holt and Roy 2007; Cáceres et al. 2009; Hawlena et al. 2010).
Can we predict whether a predator should inhibit or catalyze the spread and prevalence of disease? Ecological theory highlights both density- and trait-mediated effects of predators on the host-parasite interaction (Abrams et al. 1996; Werner and Peacor 2003; Raffel et al. 2010). Thus far, density-mediated effects dominate models for disease (e.g., the culling mechanisms described above). However, predators could indirectly influence disease by altering the traits that govern host-parasite interactions. Trait-mediated indirect effects (TMIEs) arise in a diverse array of predator–prey systems (Schmitz and Suttle 2001; Werner and Peacor 2003; Schmitz 2008). In some of these predator–prey systems, TMIEs can act very strongly (Schmitz et al. 1997; Peacor and Werner 2001; Peckarsky et al. 2008). Thus, predator-mediated TMIEs might influence host-parasite interactions, perhaps quite strongly as well (Thiemann and Wassersug 2000; Decaestecker et al. 2002; Ramirez and Snyder 2009; Raffel et al. 2010). Therefore, we need to mechanistically explain and predict predator-mediated TMIE in disease systems.
We developed a size-based mechanism for predator-mediated TMIE in a planktonic system with two types of predators. Previous work showed how vertebrate (fish) predation can reduce prevalence of fungal disease (Metschnikowia bicuspidata) in a zooplankton host (Daphnia dentifera), particularly through selective culling (Duffy et al. 2005; Duffy and Hall 2008; Hall et al. 2010b). In contrast, an invertebrate predator (Chaoborus spp.) promotes fungal epidemics (more Chaoborus, more disease: Cáceres et al. 2009; Overholt et al. 2012). It can spread disease by distributing spores while feeding (a density-mediated effect). Additionally, Chaoborus can also spread disease through TMIEs on two key traits (infection risk and fungal spore yield from infected hosts; Duffy et al. 2011). Both per capita traits increase in the presence of kairomones released from Chaoborus; we hypothesized these responses stemmed from an increase in body size of hosts (Stibor and Lüning 1994; Duffy et al. 2011).
Based on those results, here we developed a size-based model for predator-induced TMIEs. The model integrates three components. First, we identified per capita traits involved in disease spread. These are: birth and death rates of hosts; transmission rate (β), the product of exposure and susceptibility to fungal spores, hereafter called “infection risk”; feeding rate, the rate of spore contact and removal from the infectious pool (since hosts eat spores); and spore yield from dead, infected hosts (Hall et al. 2006, 2009c). Second, we mechanistically linked these focal traits to body size of hosts. Larger hosts can have higher maximal birth rates (through higher assimilation rate and more room to brood offspring: Kooijman 1993; Hall et al. 2009c). Additionally, larger hosts consume and remove more fungal spores from their environment, thereby conferring higher infection risk through enhanced exposure (Hall et al. 2007). Furthermore, larger hosts tend to yield more spores (due to energetic reasons: Hall et al. 2009b, c, 2010a, 2012). Third, we linked changes in body size of hosts to kairomones released from predators (Boersma et al. 1998; Lass and Bittner 2002; Sell 2000). Kairomones from Chaoborus often increase body size (Tollrian 1993; Weber and Declerck 1997; Sell 2000); if so, Chaoborus-induced TMIE should increase infection risk and spore yield (Duffy et al. 2011), thereby increasing disease. Kairomones from fishes typically induce smaller body size (Machacek 1995; Reede 1995; Weber and Declerck 1997; Boersma et al. 1998; Lass and Bittner 2002; Sakwinska 2002; Hesse et al. 2012). Thus, infection risk, spore yield, and birth rate should decrease due to fish-induced TMIE, thereby inhibiting disease.
We then tested predictions of this kairomone-induced size model. To synthesize key traits involved, we used a mathematical model to describe the underlying epidemiology of the Daphnia-fungus system. The model depends on the per capita traits described above (and others in Table 1). Given those traits, the model delineates an invasion criterion for the parasite, framed in terms of net reproductive ratio (R 0) (Anderson and May 1991; Hall et al. 2006, 2009c). Then, we conducted experiments to parameterize size-dependent models for the focal traits and the synthetic R 0 in no-exposure (control) or kairomone treatments (Chaoborus or fish) for six Daphnia clones. Although hosts did not respond to Chaoborus kairomones, they did grow to a smaller size with fish kairomones, thereby depressing several key traits involved in disease spread (especially birth rate and spore yield). However, the infectivity trait contradicted predictions of the size model because fish kairomones increased per spore susceptibility, another component of infection risk (Yin et al. 2011; Civitello et al. 2012, 2013). This tension between size-based traits (decreasing R 0) and susceptibility (increasing R 0) yielded no net effect of fish kairomones on R 0.
Materials and methods
Mathematical model
We used a mathematical model to create testable predictions and to synthesize parameter estimates (from experiments) into a single disease metric. The model is built around the epidemiology of parasites that produce free-living dispersal stages upon host death (i.e., “obligate killers”; Hall et al. 2006; Cáceres et al. 2009). It tracks changes in susceptible host density (S), infected host density (I), and free-living infective stages of the parasite (Z):
The first equation (Eq. 1.a) tracks change in S through time as a function of births [with maximal birth rates for susceptible (b) and infected hosts (b I ), respectively, which are then depressed by a density-dependence parameter (c)]; deaths [at background rate (d)]; and disease transmission following a density-dependent (mass action) interaction with infection risk (transmission rate; β). We build a more mechanistic representation of β below. I (Eq. 1.b) increases as susceptible hosts become infected (βSZ), but die at a rate elevated by infection (d + v). Finally, free-living infective stages [spores (Z); Eq. 1.c] increase as infected hosts die and release spores, with per capita spore yield (σ). They then decrease at background loss rate m and through consumption by hosts (S + I) which feed at rate f.
Using this model, we can calculate a key disease metric, net reproductive ratio (R 0). This metric determines the ability of a parasite to invade (when R 0 > 1) and typically scales proportionally to equilibrium infection prevalence. For this model (Eq. 1), R 0 is:
The first term in parentheses contains birth rate (b) and death rate (d) of susceptible hosts, two traits estimated in our experiments. If kairomones decrease b or increase d, they will inhibit epidemics. [We did not measure c, but stronger density dependence on b (higher c) decreases R 0.] The second term in parentheses shows that epidemics are inhibited if kairomones depress spore yield from dead, infected hosts (σ) or infection risk (β). At first glance, R 0 decreases with feeding rate (f). However, β is really the product of exposure due to feeding, f and the per spore susceptibility of hosts to consumed spores (u): β = uf. With this mechanism, R 0 decreases if f decreases (provided that σu > 1) and/or u decreases. Thus, kairomones can inhibit disease if they decrease b, σ, exposure (f), and/or increase d. However, they can increase it by elevating u.
In a series of experiments, we estimate these key parameters (b, background d, σ, f, u). Although we explain details of those experiments below, we can mechanistically connect kairomones, body size, and two parameters: σ and β. σ commonly increases with body size of hosts upon death (L d ) (Hall et al. 2009b, c; Duffy et al. 2011). A simple model for σ then is
which says that σ should increase linearly with size at death (with slope σ1 and intercept σ0). Thus, kairomones can depress σ if hosts die at smaller size (L d ). β increases with body size at time of infection (L β) in part because exposure (feeding rate; f) increases with body size. More specifically, f should increase with surface area, or \( L_{\beta }^{2} \), \( f = \hat{f}L_{\beta }^{2} \) (where \( \hat{f} \) is size-specific (size-controlled) f; Hall et al. 2007). β increases even more steeply with body size (statistically, with a length4 relationship: Hall et al. 2007). Thus, if we add an additional size factor \( \left( {L_{\beta }^{2} } \right) \), we can more mechanistically represent β as:
This model means that kairomones could alter β through effects on L β, size-corrected f (\( \hat{f} \)), and/or u. For instance, kairomones could depress body size but net increase β by dramatically increasing per spore susceptibility. The experiments below delineate among these three components.
Experiments
We combined three experiments to estimate key parameters (b, d, σ, f, u) involved in the model. All experiments involved six genotypes of Daphnia dentifera, collected from three lakes (see Table A1), raised in one of three treatment waters: kairomones of bluegill sunfish (Lepomis macrochirus), kairomones of larvae of the phantom midge Chaoborus punctipennis, or a predator-free control. Daphnia genotypes were collected from lakes in southwest Michigan (Barry County), and are known to vary in their susceptibility to Metschnikowia (Duffy and Sivars-Becker 2007; Hall et al. 2010a). The parasite was collected from Baker Lake in 2003 and has been farmed in vivo in a single host clone (Standard). Unlike in other Daphnia disease systems (Ebert 2005), infection does not depend on an interaction between host and parasite genotypes (i.e., there is no genetic specificity in the D. dentifera-Metschnikowia system; Duffy and Sivars-Becker 2007; Searle et al., in preparation). The predators were collected from a local lake in Illinois.
To create media with kairomones, we filled two replicate 19-L aquaria for each treatment with filtered aged lake water. Chaoborus (third to fourth instars) tanks contained ~8 Chaoborus/L, replaced when needed, and fed laboratory cultured live Daphnia ad libitum. To control for kairomones that could potentially be released from Daphnia, laboratory-cultured live Daphnia were also introduced initially into the control tanks and fish tanks. Each fish tank contained from four to five bluegill [total mass, 19.6 ± 3.0 g (mean ± SE)], supplemented with 2.5 g frozen Daphnia/tank per day (Bio-pure Daphnia; Hikari Aquatic Diets, Hayward, CA). Prior to use, culture media was sieved twice through a 30-μm mesh.
To minimize maternal effects, all experimental animals were collected from the third or later clutch, following at least three generations of acclimation to control conditions. During this acclimation period, cultures were maintained at low density in 20 °C on a 14:10-h light:dark cycle and fed 40,000 cells/ml per day of Ankistrodesmus falcatus (a green alga).
b and d
To parameterize the life history components of our model we used a life-table protocol modified from Lynch et al. (1989). Newborn D. dentifera from six clonal lines were placed individually in 150-ml beakers containing 110 ml of treatment media (control, Chaoborus or fish, n = 10 of each genotype per treatment). Each animal was moved to fresh treatment water with food (40,000 cells/ml Ankistrodesmus) every other day. Cultures were maintained at 20 °C on a 14:10-h light:dark cycle and Daphnia were checked daily for maturation and reproduction for 40 days. Age and size were recorded at the production of the first clutch. Details of how b and d were estimated from these data are provided in the Electronic supplementary material in the Appendix.
f assays
To determine if and how predators affect D. dentifera feeding rate, we conducted an assay using control, Chaoborus, and fish treatments. Experimental animals were reared as described for the previous assays. When 6 days old, five Daphnia from each genotype × treatment combination were placed individually in 50-ml centrifuge tubes containing 45 ml of treatment media. After a 2-h acclimation period, they were inoculated with 40,000 cells Ankistrodesmus/ml and allowed to feed for 24 h. Ten identical tubes without animals were also inoculated, with half being preserved using Lugol’s iodine solution immediately and the other half being preserved at the end of the experiment. This provided the initial cell concentration with adjustment for reproduction/degradation of algae over the 24-h period. After 24 h, Daphnia were removed from the tubes, preserved in 95 % ethanol, and subsequently measured (top of the head to base of the tail spine). Algal cells were concentrated and density was determined using a hemocytometer.
Estimating infection components (β, f, u, σ)
Infection risk, β, exposure (feeding) rate (f), and per spore susceptibility, u were determined by combining fits of models to infection data and feeding rate data described above. For details on model fitting, see Electronic supplementary material in the Appendix. Infection and feeding rate data were collected in separate experiments but combined in the model fitting. To quantify infection and spore yield, six to eight replicates of each of the Daphnia genotypes were raised in one of three treatment waters from birth. Eight newborn Daphnia were placed in 150-ml beakers containing 110 ml of treatment media. After 8 days, beakers were culled to six Daphnia, inoculated with 100 Metschnikowia spores/ml, and the Daphnia were measured to determine size at inoculation. The following day, all Daphnia were moved to fresh media and fed 40,000 cells/ml Ankistrodesmus. Infection was visually assessed using a dissecting microscope 10 days following inoculation. Spore yield was estimated by haphazardly selecting one infected animal from each beaker (resulting in up to eight infected animals per genotype per treatment). Individuals were put into 24-well tissue culture plates until they succumbed to infection. These infected animals were measured to determine size at death and homogenized in 1 ml of water to release all spores from the body cavity. Spore yield was determined using a hemocytometer.
Statistical analysis
We used ANOVA in Systat 13 to determine the effect of exposure to fish or Chaoborus kairomones on the various components of R 0 (b, d, f, β, σ). Each genotype yielded a point estimate of a parameter and R 0. All of these values were estimated using code written for Matlab. Thus, genotype was used as the unit of replication for all analyses, but we bootstrapped SEs for each R 0 estimate for each genotype (again using Matlab; see Supplementary in Appendix). See Electronic supplementary material in the Appendix for additional details on the parameter estimation.
Results
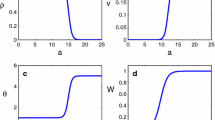
Consistent with our predictions, we found that fish kairomones depressed b (Fig. 1a) and increased d (Fig. 1b), relative to both the control and Chaoborus treatments. The fish-induced decline in b stemmed from longer time to maturity (higher age at first reproduction) and lower fecundity per day shown by those clones exposed to fish kairomones (Appendix, Figs. A1A, B). The lower fecundity was also associated with smaller body size of hosts in the presence of fish kairomones (Appendix, Fig. A1C). Hosts dying from infection were also smaller when exposed to fish kairomones (Fig. 1c). As anticipated, those smaller hosts yielded fewer spores upon death from infection (Fig. 1d, e). Thus, at this point, the size model seemed to work for fish cues. However, in all of these response variables, Chaoborus treatments responded similarly to controls (even for the body size indices). Thus, we de-emphasize size-based predictions for this treatment below.
a Birth rate (b) and b death rate (d) components of the net reproductive ratio (R 0; Eq. 2) for six clones of Daphnia dentifera exposed to three treatments: control or kairomones from two predators, Chaoborus (Chaob.) or fish (means ± 1 SE). Different letters within the bars indicate significant differences among treatments according to Tukey’s honestly significant difference (HSD) test at P < 0.05. a Instantaneous b. b Instantaneous d. c, d Spore yield components of the net reproductive ratio (R 0). c Size at death of infected hosts (L d ). d Spore yield from dead, infected hosts (σ). e Positive correlation between L d and σ (as anticipated, Eq. 3)
In the fish treatment, the size-based model falls apart for the infection risk trait. Although fish-exposed hosts were significantly smaller at infection (Fig. 2a), we found no difference among treatments in β (Fig. 2b). When controlled for size at infection, fish-exposed hosts had a significantly higher (size-specific) infection rate (Fig. 2c). If infection risk were solely influenced by the size response of hosts to kairomones, we would have expected no difference between fish and control treatments in this size-specific β. (The Chaoborus treatments did not respond differently than the controls).
Infection risk (β) components of R 0 (Eq. 2) for six clones of D. dentifera exposed to three treatments: control or kairomones from two predators, Chaoborus or fish (means ± 1SE). a Size at infection (L β). b β and c size-specific β, \( \hat{\beta } \), i.e., β when controlling for L β following Eq. 4. d Size-specific exposure risk, \( \hat{f} \), (i.e., ‘clearance rate’ in the resource consumption sense), and e exposure risk given size at infection (f). f Per spore susceptibility (u) i.e., infectivity of spores once consumed. Different letters within the bars indicate significant differences among treatments according to Tukey’s HSD test at P < 0.05. For other abbreviations, see Fig. 1
Analysis of the components of infection risk (Eq. 4) explained this response. Fish kairomones could have elevated β by increasing exposure through size-controlled feeding rate, \( \hat{f} \). In principle, higher size-controlled feeding rate could overwhelm the (lower) body-size component of exposure (L β). Our feeding rate data rule this case out (Fig. 2d, e). In the fish media, we found no significant difference among treatments in the size-specific exposure (feeding) rate, \( \hat{f} \), and reduced overall exposure (size-specific feeding rate times size at infection2: \( f = \hat{f}L_{\beta }^{2} \); Fig. 2d, e). Instead, fish influenced per spore susceptibility to parasites. Estimates of infectivity (u) were higher for hosts exposed to fish kairomones than to controls (but not Chaoborus kairomones: Fig. 2f). This susceptibility trait thus pulled in a direction opposite to that of the birth and spore yield traits in fish media (Fig. 1).
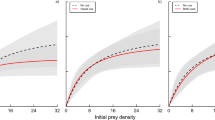
Tension between these traits then yielded no net effect of fish kairomones on the synthetic metric of disease spread (R 0). Hosts exposed to kairomones from fish yielded similar R 0 estimates as those in the control or the Chaoborus treatments (Fig. 3a). When combined, the uninfected b and d rate components of R 0 decreased in the treatment with fish kairomones (as expected; Fig. 3b). However, the net product of spore yield and infection risk, once spore removal (f) was accounted for (i.e., σβ−f), yielded no differences among treatments but high variance among point estimates for the six host genotypes (Fig. 3c). This high variability stems from positive correlations between σ and β among clones (Figure A2A): clones with high β produced more spores. This variance all but eliminates net differences in R 0 among kairomone treatments. Clones vary in their R 0 values (i.e., some clones are more apt to spread disease than others), but not in an interactive manner with treatment (Fig. A2B).
R 0 (Eq. 2), integrating b and d, σ, β, and f for six clones of D. dentifera exposed to control or kairomones from two predators, Chaoborus or fish (means ± 1 SE). a Net reproductive ratio (R 0). The culmination of b uninfected components [(b−d)/b], and c infected components (σβ−f). Other parameters in R 0 calculation: density-dependence of b (c = 0.1 L host−1); background loss rate of spores (m = 0.5 day−1). Both c and m are set at reasonable rates for this system, but the qualitative conclusions regarding treatment differences/similarities do not depend on them. Different letters within the bars indicate significant differences among treatments according to Tukey’s HSD test at P < 0.05. For other abbreviations, see Figs. 1 and 2
Discussion
We tested a mechanistic model that predicted TMIEs on core components of disease spread in a focal zooplankton host fungal parasite system. Predators can alter disease through these TMIEs on host-prey behavior (Thiemann and Wassersug 2000; Decaestecker et al. 2002; Daly and Johnson 2011) and immune function (Rigby and Jokela 2000; Coslovsky and Richner 2011). Here, we looked for them through kairomone effects on host physiology and a resulting shift in the response of life history traits (Rinke et al. 2008; Duffy et al. 2011; Yin et al. 2011). Previous work in this host-parasite system showed how the size-based response of hosts to kairomones from an invertebrate predator increased body size of hosts (Duffy et al. 2011). This predation defense reflects allocation of energy to growth rather than reproduction (Stibor and Lüning 1994; Reissen 1999; Rinke et al. 2008; but see Noonburg and Nisbet 2005). This body size response to Chaoborus then elevated infection risk and spore yield from infected hosts (Duffy et al. 2011), as anticipated from size-based models of exposure and energetics (Hall et al. 2007, 2009c, 2010a). Here, we extended that size-based model for predator-induced TMIE to a fish, an important predator of the host.
Several factors responded as anticipated for the treatment with fish kairomones. First, body size was smaller at time of infection (8 days old). This result reflects a strategy to avoid predation from size-selective fish predators (Stibor and Lüning 1994; Boersma et al. 1998). This size effect, and perhaps also a predator—induced stress response (unmeasured, but seen in other systems; e.g., Boonstra et al. 1998; Hawlena and Schmitz 2010; McCauley et al. 2011), translated into lower instantaneous birth rates (b in the dynamical model) and elevated age at first reproduction. This latter finding contrasts with those from a similar study involving Daphnia hosts exposed to both fish and Metschnikowia (Yin et al. 2011). Perhaps age at first reproduction depends on kairomone-induced changes in allocation, stress, energy investment per offspring, etc., in a manner that could produce either outcome [Kooijman 1993; see Appendix of Duffy et al. (2011) for some modeling results pointing to this possibility]. Regardless, d of hosts exposed to fish kairomones also increased. Furthermore, spore yield dropped with fish cues—as expected from size-based energetic models (Hall et al. 2009c, Duffy et al. 2011) but opposite that in another study (Yin et al. 2011). This drop in σ matters because disease spread depends on spore densities, yet spores may suffer high loss rates [due to damage from solar radiation (Overholt et al. 2012) and/or consumption by other hosts (Hall et al. 2009a)]. When combined, these traits point to strong inhibitory effects of fish kairomones on disease spread and confirm the underlying size model.
The infection risk trait, however, contradicted the size model. Since fish kairomones reduced body size of hosts, it should have reduced infection risk, all else equal, through lowered exposure (Hall et al. 2007; Duffy et al. 2011). Instead, β did not change significantly among treatments. Why should smaller hosts have higher size-controlled vulnerability to infection? Our data dispelled size-specific feeding rate as the answer. Very fast size-specific feeding, in principle, could have overcome smaller body size to elevate overall exposure. Instead, per spore susceptibility increased with fish kairomones. At this point, we cannot pinpoint causation for the boosted susceptibility, but stress from the predator cues might weaken the immune response of hosts (Pauwels et al. 2010) or perhaps change some aspect of the host gut, the last barrier between fungal spore and hosts (Rohrlack et al. 2005; Dussaubat et al. 2012). This response of susceptibility to fish kairomones, however, yielded no net difference for both β and the R 0 among treatments. Thus, susceptibility swamped out exposure and other size-based traits responding to fish kairomones.
We have seen other examples of this tension between components of disease traits before in this system. For instance, resource quality and quantity can elevate birth rates and spore yield from infected hosts, via energetic mechanisms, while decreasing β, via contact mechanisms (Hall et al. 2007, 2009b, c). Similarly, copper contamination can decrease spore yield from infected hosts (presumably through toxic effects on energetics) while elevating infection risk (through boosted feeding rate: Civitello et al. 2012). These previous examples, coupled with the present case with kairomones, indicate that environmental gradients can inhibit, elevate, or neutrally affect disease depending on net outcome of the competition between responses of traits involved. In particular, the net outcome depends on whether the environmental influence on spore yield can surmount that on infection risk. Here, these factors essentially cancel each other out. That conclusion might change in other environmental contexts.
The kairomone treatments using the invertebrate predator Chaoborus did not recapture results from a previous study (Duffy et al. 2011). Here, individual traits and R 0 differed little between control and Chaoborus kairomone treatments. Point estimates for R 0 were higher for four of six host clones involved, but error propagation among traits yielded too much uncertainty to conclude much more. This point emphasizes the high replication demands of estimating compound parameters like R 0. The squelched response to kairomones of Chaoborus could reflect, however, the experimental conditions. Hosts in better conditions (like in this experiment) often respond less to kairomones than hosts experiencing somewhat worse conditions (like in the sister study: Duffy et al. 2011; Pijanowska et al. 2006; Pauwels et al. 2010). However, confirming such an idea would require additional experiments.
In the meantime, we can interpret these results through both a narrower and broader lens. A narrow reading points to no strong net effect of fish kairomones on disease spread. Thus, the pattern detected in field surveys (smaller epidemics with more intense fish predation: Duffy et al. 2005; Johnson et al. 2006; Hall et al. 2010b) likely stems from density-mediated rather than trait-mediated mechanisms of fish. A broader interpretation highlights two core results and emphasizes a question for the future. First, the size-based model for predator-induced TMIE worked nicely for two traits (birth rate and spore yield). This means that the physiological response of hosts to predator kairomones can predict effects on at least some disease traits. Second, predator-induced TMIE can transcend these size-based mechanisms to potently influence susceptibility. The underlying susceptibility response, driven by mechanisms unexplored here, can rival those produced by the size-exposure component of β alone. This result then points to the need to synthesize physiological, stress, anatomical, and immune responses when predicting predator-induced TMIE on host-parasite interactions.
References
Abrams P, Menge BA, Mittelbach GG, Spiller D, Yodzis P (1996) The role of indirect effects in food webs. In: Polis GA, Winemiller KO (eds) Food webs: integration of patterns and dynamic. Chapman and Hall, New York, pp 371–395
Anderson RM, May RM (1991) Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford
Boersma M, Spaak P, De Meester L (1998) Predator-mediated plasticity in morphology, life history, and behavior in Daphnia: the uncoupling of responses. Am Nat 152:237–248
Boonstra R, Hik D, Singleton GR, Tinnikov A (1998) The impact of predator-induced stress on the snowshoe hare cycle. Ecol Monogr 79:371–394
Cáceres CE, Knight CJ, Hall SR (2009) Predator-spreaders: predation can enhance parasite success in a planktonic host-parasite system. Ecology 90:2850–2858
Choisy M, Rohani P (2006) Harvesting can increase severity of wildlife disease epidemics. Proc R Soc B Biol Sci 273:2025–2034
Civitello DJ, Forys P, Johnson AP, Hall SR (2012) Chronic contamination decreases disease spread: a Daphnia-fungus-copper case study. Proc R Soc B Biol Sci 279:3146–3153
Civitello DJ, Penczykowski RM, Hite JL, Duffy MA, Hall SR (2013) Potassium stimulates fungal epidemics in Daphnia by increasing host and parasite reproduction. Ecology 94:380–388
Coslovsky M, Richner H (2011) Predation risk affects offspring growth via maternal effects. Funct Ecol 25:878–888
Daly EW, Johnson PTJ (2011) Beyond immunity: quantifying the effects of host anti-parasite behavior on parasite transmission. Oecologia 165:1043–1050
Daszak P, Cunningham AA, Hyatt AD (2000) Wildlife ecology: emerging infectious diseases of wildlife—threats to biodiversity and human health. Science 287:443–449
Decaestecker E, De Meester L, Ebert D (2002) In deep trouble: habitat selection constrained by multiple enemies in zooplankton. Proc Natl Acad Sci 99:5481–5485
Dobson AP, Foufopoulos J (2001) Emerging infectious pathogens in wildlife. Philos Trans R Soc Lond B 356:1001–1012
Duffy MA, Hall SR (2008) Selective predation and rapid evolution can jointly dampen effects of virulent parasites on Daphnia populations. Am Nat 171:499–510
Duffy MA, Sivars-Becker L (2007) Rapid evolution and ecological host-parasite dynamics. Ecol Lett 10:44–53
Duffy MA, Hall SR, Tessier AJ, Huebner M (2005) Selective predators and their parasitized prey: are epidemics in zooplankton under top-down control? Limnol Oceanogr 50:412–420
Duffy MA, Housley JM, Penczykowski RM, Cáceres CE, Hall SR (2011) Unhealthy herds: indirect effects of predators enhance two drivers of disease spread. Funct Ecol 25:945–953
Dussaubat C, Brunet J-L, Higes M, Colbourne JK, Lopez J, Choi J-H, Martín-Hernádez R, Botías C, Cousin M, McDonnell C, Bonnet M, Belzunces LP, Moritz RF, Le Conte Y, Alaux C (2012) Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS One 7(5):e37017. doi:10.1371/journal.pone.0037017
Ebert D (2005) Ecology, epidemiology, and evolution of parasitism in Daphnia [Internet]. National Center for Biotechnology Information, National Library of Medicine (US), Bethesda, MD. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
Hall SR, Duffy MA, Cáceres CE (2005) Selective predation and productivity jointly drive complex behavior in host-parasite systems. Am Nat 165:70–81
Hall SR, Tessier AJ, Duffy MA, Huebner M, Cáceres CE (2006) Warmer does not have to mean sicker: temperature and predators can jointly drive timing of epidemics. Ecology 87:1684–1695
Hall SR, Sivars-Becker L, Becker C, Duffy MA, Tessier AJ, Cáceres CE (2007) Eating yourself sick: transmission of disease as a function of foraging ecology. Ecol Lett 10:207–218
Hall SR, Becker CR, Simonis JL, Duffy MA, Tessier AJ, Cáceres CE (2009a) Friendly competition: evidence for a dilution effect among competitors in a planktonic host-parasite system. Ecology 90:791–801
Hall SR, Knight CJ, Becker CR, Duffy MA, Tessier AJ, Cáceres CE (2009b) Quality matters: resource quality for hosts and the timing of epidemics. Ecol Lett 12:118–128
Hall SR, Simonis JL, Nisbet RM, Tessier AJ, Cáceres CE (2009c) Resource ecology of virulence in a planktonic host-parasite system: an explanation using dynamic energy budgets. Am Nat 174:149–162
Hall SR, Becker C, Duffy MA, Cáceres CE (2010a) Variation in resource acquisition and use among host clones creates key epidemiological trade-offs. Am Nat 176:557–565
Hall SR, Smyth R, Becker CR, Duffy MA, Knight CJ, MacIntyre S, Tessier AJ, Cáceres CE (2010b) Why are Daphnia in some lakes sicker? Disease ecology, habitat structure, and the plankton. Bioscience 60:363–375
Hall SR, Becker C, Duffy MA, Cáceres CE (2012) A power-efficiency tradeoff alters epidemiological relationships. Ecology 93:645–656
Harvell CD, Mitchell CE, Ward JR, Altizer S, Dobson AP, Ostfeld RS, Samuel MD (1999) Emerging marine diseases—climate links and anthropogenic factors. Science 285:1505–1510
Hatcher MJ, Dick JT, Dunn AM (2006) How parasites affect interactions between competitors and predators. Ecol Lett 9:1–19
Hawlena D, Schmitz OJ (2010) Physiological stress as a fundamental mechanism linking predation to ecosystem function. Am Nat 176:537–556
Hawlena D, Abramsky Z, Bouskila A (2010) Bird predation alters infestation of desert lizards by parasitic mites. Oikos 119:730–736
Hesse O, Engelbrecht W, Laforsch C, Wolinska J (2012) Fighting parasites and predators: how to deal with multiple threats? BMC Ecol 12:12. doi:10.1186/1472-6785-12-12
Holt RD, Roy M (2007) Predation can increase the prevalence of infectious disease. Am Nat 169:690–699
Johnson PTJ, Stanton DE, Preu ER, Forshay KJ, Carpenter SR (2006) Dining on disease: how interactions between infection and environment affect predation risk. Ecology 87:1973–1980
Keesing F, Holt RD, Ostfeld RS (2006) Effects of species diversity on disease risk. Ecol Lett 9:485–498
Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, Hudson P, Jolles A, Jones KE, Mitchell CE, Myers SS, Bogich T, Ostfeld RS (2010) Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468:647–652
Kilpatrick AM (2011) Globalization, land use, and the emergence of West Nile virus. Science 334:323–327
Kooijman SALM (1993) Dynamic energy budgets in biological systems. Cambridge University Press, New York
Lass S, Bittner K (2002) Facing multiple enemies: parasitised hosts respond to predator kairomones. Oecologia 132:344–349
Lynch M, Spitze K, Crease T (1989) The distribution of life-history variation in the Daphnia pulex complex. Evolution 43:1724–1736
Machacek J (1995) Inducibility of life-history changes by fish kairomone in various developmental stages of Daphnia. J Plankton Res 17:1513–1520
McCauley SJ, Rowe L, Fortin M-J (2011) The deadly effects of “nonlethal” predators. Ecology 92:2043–2048
Noonburg EG, Nisbet RM (2005) Behavioral and physiological responses to food availability and predation risk. Evol Ecol Res 7:89–104
Ostfeld RS, Holt RD (2004) Are predators good for your health? Evaluating evidence for top-down regulation of zoonotic disease reservoirs. Front Ecol Environ 2:13–20
Overholt EP, Hall SR, Williamson CE, Meikle CK, Duffy MA, Cáceres CE (2012) Solar radiation decreases parasitism in Daphnia. Ecol Lett 15:47–54
Packer C, Holt RD, Hudson PJ, Lafferty KD, Dobson AP (2003) Keeping the herds healthy and alert: implications of predator control for infectious disease. Ecol Lett 6:797–802
Pauwels K, Stoks R, De Meester L (2010) Enhanced anti-predator defense in the presence of food stress in the water flea Daphnia magna. Funct Ecol 24:322–329
Peacor SD, Werner EE (2001) The contribution of trait-mediated indirect effects to the net effects of a predator. Proc Natl Acad Sci 98:3904–3908
Peckarsky BL, Abrams PA, Bolnick DI, Dill LM, Grabowski JH, Luttbeg B, Orrock JL, Peacor SD, Preisser EL, Schmitz OJ, Trussell GC (2008) Revisiting the classics: considering nonconsumptive effects in textbook examples of predator-prey interactions. Ecology 89:2416–2425
Pijanowska J, Dawidowicz P, Howe A, Weider LJ (2006) Predator induced shifts in Daphnia life-histories under different food regimes. Arch Hydrobiol 167:37–54
Raffel TR, Hoverman JT, Halstead NT, Michel PJ, Rohr JR (2010) Parasitism in a community context: trait-mediated interactions with competition and predation. Ecology 91:1900–1907
Ramirez RA, Snyder WE (2009) Scared sick? Predator-pathogen facilitation enhances exploitation of a shared resource. Ecology 90:2832–2839
Reede T (1995) Life history shifts in response to different levels of fish kairomones in Daphnia. J Plankton Res 17:1661–1667
Reissen HP (1999) Chaoborus predation and delayed reproduction in Daphnia: a demographic modeling approach. Evol Ecol 13:339–363
Rigby MC, Jokela J (2000) Predator avoidance and immune defence: costs and trade-offs in snails. Proc R Soc Lond B 267:171–176
Rinke K, Hulsmann S, Mooij WM (2008) Energetic costs, underlying resource allocation patterns, and adaptive value of predator-induced life-history shifts. Oikos 117:273–285
Rohrlack KC, Dittmann E, Norgueira I, Vasconcelos V, Börner T (2005) Ingestion of microcystins by Daphnia: intestinal uptake and toxic effects. Limnol Oceanogr 50:440–448
Sakwinska O (2002) Response to fish kairomone in Daphnia galeata life history traits relies on shift to earlier instar at maturation. Oecologia 131:409–417
Schmitz OJ (2008) Effects of predator hunting mode on grassland ecosystem function. Science 319:952–954
Schmitz OJ, Suttle KB (2001) Effects of top predator species on direct and indirect interactions in a food web. Ecology 82:2072–2081
Schmitz OJ, Beckerman AP, O’Brian KM (1997) Behaviorally-mediated trophic cascades: the effects of predation risk on food web interactions. Ecology 78:1388–1399
Sell AF (2000) Morphological defenses induced in situ by the invertebrate predator Chaoborus: comparison of responses between Daphnia pulex and D. rosea. Oecologia 125:150–160
Stibor H, Lüning J (1994) Predator-induced phenotypic variation in the pattern of growth and reproduction in Daphnia hyalina (Crustacea, Cladocera). Funct Ecol 8:98–101
Thiemann GW, Wassersug RJ (2000) Patterns and consequences of behavioural responses to predators and parasites in Rana tadpoles. Biol J Linn Soc 71:513–528
Tollrian R (1993) Neckteeth formation in Daphnia pulex as an example of continuous phenotypic plasticity: morphological effects of Chaoborus kairomone concentration and their quantification. J Plankton Res 15:1309–1318
Weber A, Declerck S (1997) Phenotypic plasticity of Daphnia life history traits in response to predator kairomones: genetic variability and evolutionary potential. Hydrobiologia 360:89–99
Werner EE, Peacor SD (2003) A review of trait-mediated indirect interactions in ecological communities. Ecology 84:1083–1100
Yin M, Laforsch C, Lohr J, Wolinska J (2011) Predator-induced defense makes Daphnia more vulnerable to parasites. Evolution 65:1482–1488
Acknowledgments
This research was supported by National Science Foundation grants DEB 0613510, 0614316, 1120804, and 1120316.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Steven Kohler.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bertram, C.R., Pinkowski, M., Hall, S.R. et al. Trait-mediated indirect effects, predators, and disease: test of a size-based model. Oecologia 173, 1023–1032 (2013). https://doi.org/10.1007/s00442-013-2673-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-013-2673-0