Abstract
Mutualisms with facultative, non-essential heritable microorganisms influence the biology of many insects, and they can have major effects on insect host fitness in certain situations. One of the best-known examples is found in aphids where the facultative endosymbiotic bacterium Hamiltonella defensa confers protection against hymenopterous parasitoids. This symbiont is widely distributed in aphids and related insects, yet its defensive properties have only been tested in two aphid species. In a wild population of the grain aphid, Sitobion avenae, we identified several distinct strains of endosymbiotic bacteria, including Hamiltonella. The symbiont had no consistent effect on grain aphid fecundity, though we did find a significant interaction between aphid genotype by symbiont status. In contrast to findings in other aphid species, Hamiltonella did not reduce aphid susceptibility to two species of parasitoids (Aphidius ervi and Ephedrus plagiator), nor did it affect the fitness of wasps that successfully completed development. Despite this, experienced females of both parasitoid species preferentially oviposited into uninfected hosts when given a choice between genetically identical individuals with or without Hamiltonella. Thus, although Hamiltonella does not always increase resistance to parasitism, it may reduce the risk of parasitism in its aphid hosts by making them less attractive to searching parasitoids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most of the world’s insect species host non-essential endosymbiotic bacteria within their tissues and cells (Hilgenboecker et al. 2008; Moran et al. 2008). These heritable microorganisms have long been primarily regarded as reproductive parasites, manipulating their hosts’ reproduction in ways that facilitate their spread within populations (Duron et al. 2008; Engelstadter and Hurst 2009). However, in the last decade it has become clear that many of these facultative symbionts confer benefits to their hosts, including resistance to natural enemies and abiotic stressors, or improved performance on certain diets (Himler et al. 2011; Łukasik et al. 2013; Montllor et al. 2002; Oliver et al. 2003; Tsuchida et al. 2004). The extent to which these benefits are common features of insect endosymbiosis will have important effects on the structure and evolution of their hosts’ populations and communities (Feldhaar 2011; Ferrari and Vavre 2011; Oliver et al. 2010). However, even though the number of studied systems has recently increased, there is still relatively little information on the diversity and roles of facultative endosymbionts outside of a few model organisms. Here, we investigate the effects of one of the best studied facultative symbionts of insects, a gammaproteobacterium Hamiltonella defensa (Moran et al. 2005b), on the grain aphid Sitobion avenae. S. avenae is a member of the same tribe (Macrosiphini) as the pea aphid, Acyrthosiphon pisum, in which Hamiltonella and other facultative endosymbionts have been most comprehensively studied (Oliver et al. 2010).
A pea aphid individual can host one or more of at least eight known genera of facultative endosymbionts (Oliver et al. 2010; Russell et al. 2013), most of which have also been reported from other species of aphids and in some cases more distantly related insects (Burke et al. 2009; Fukatsu et al. 2001; Russell et al. 2003; Sandstrom et al. 2001). The prevalence of these bacteria in populations varies considerably across pea aphid host plant races and geographic areas (Ferrari et al. 2012; Russell et al. 2013; Tsuchida et al. 2002), possibly influenced by the selective advantage of carrying different symbionts in different habitats (Oliver et al. 2008, 2010). Most, if not all, of the known pea aphid facultative endosymbionts are capable of conferring protection against natural enemies, including pathogens (Łukasik et al. 2013; Scarborough et al. 2005) and parasitoid wasps (Guay et al. 2009; Nyabuga et al. 2010; Oliver et al. 2003). Facultative endosymbionts can also influence pea aphid susceptibility to heat shock (Montllor et al. 2002; Russell and Moran 2006), affect their mode of reproduction (Leonardo and Mondor 2006), alter performance across host plants (McLean et al. 2011; Tsuchida et al. 2004), or influence body coloration (Tsuchida et al. 2010).
Hamiltonella defensa is one of the most common symbionts in A. pisum populations (Ferrari et al. 2012) and protects pea aphids against hymenopterous parasitoids (Oliver et al. 2003). It has the same effect in the black bean aphid, Aphis fabae (Schmid et al. 2012) and there is evidence that it can also protect the cowpea aphid, Aphis craccivora (Desneux et al. 2009). Strains of Hamiltonella from the pea aphid differ in the degree of protection they confer against the parasitoid Aphidius ervi, which is reflected in both the survival and fecundity of parasitized aphids (Oliver et al. 2005). These differences are linked to infection with a lysogenic lambdoid bacteriophage called Acyrthosiphon pisum secondary endosymbiont (after its host, APSE) that encodes one of several distinct eukaryotic toxins (Degnan and Moran 2008a, b; Moran et al. 2005a; Oliver et al. 2009). Different Hamiltonella-APSE associations vary in their effects on parasitoid survival (Oliver et al. 2005), and parasitoid genotypes differ in their response to symbiont-conferred protection (Dion et al. 2011; Schmid et al. 2012). Parasitoids are capable of detecting the infection status of their prospective pea aphid hosts and can respond by preferentially superparasitizing symbiont-protected aphids, which helps them to overcome their defences (Oliver et al. 2012).
Though we now know a considerable amount about the endosymbionts of pea aphids and to a lesser extent black bean aphids, our knowledge of Hamiltonella and other facultative endosymbionts in further aphid species is typically limited to surveys involving one or few individuals (Burke et al. 2009; Haynes et al. 2003; Russell et al. 2003; Sandstrom et al. 2001). There is little information about strain diversity or the effects symbionts have on these hosts (Chen et al. 2000; Łukasik et al. 2011). In this study, we assessed the diversity of facultative endosymbionts in an English population of the grain aphid, Sitobion avenae, one of the world’s most serious pests of cereals (Dedryver et al. 2010). We then tested the hypothesis that Hamiltonella confers similar benefits to S. avenae as it does to the closely related A. pisum. We introduced bacteria into naturally facultative symbiont-free aphid genotypes, or into lines previously cured from infection, to measure the effects symbionts have on aphid fecundity and susceptibility to parasitoids. We predicted that Hamiltonella would increase resistance to two species of aphidiine parasitoids that commonly attack grain aphids in England, A. ervi and Ephedrus plagiator (Muller et al. 1999; Powell 1982; Traugott et al. 2008). We also studied the development times and sizes of parasitoids successfully completing development in infected and uninfected hosts and asked whether parasitoids discriminate between these two types of host at oviposition as they do in pea aphids (Oliver et al. 2012).
Materials and methods
Aphids and their facultative endosymbionts
S. avenae were collected in June 2008 from wheat (Triticum sp.) and cocksfoot grass (Dactylis glomerata) near Great Coxwell, Oxfordshire, UK, and from oat (Avena sativa) and Dactylis near Lower Radley, Oxfordshire, UK, approximately 30 km east of the first site. To reduce the risk of sampling the same genotype multiple times, individual aphids were collected from plants growing at least 5 m apart. After collection, clonal lines were cultured in 90-mm non-vented Petri dishes on approximately 10-day-old Triticum plants that had their stems placed in 2 % agar. Plants were renewed approximately every 10 days. The conditions in the culture room were 14 ± 1 °C and a 16:8-h light:dark regime, ensuring indefinite asexual reproduction of aphids. Before any experiments, the insects were kept at 20 ± 2 °C (the temperature used during experiments) on plants renewed every 3–4 days, for at least three generations (Łukasik et al. 2011).
DNA from field-collected adult aphids that were used to establish laboratory clonal lines was extracted with DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. Species-level identification based on morphological characters was confirmed by amplification and sequencing of the mitochondrial cytochrome c oxidase 1 (COI) gene, the standard DNA barcode region for animal life (Foottit et al. 2008), for most lines, including all those later used in the experiments. Next, all aphids were typed at seven microsatellite loci (Łukasik et al. 2011; Table S1) to ensure that they were distinct genotypes. The samples were then screened for the seven facultative endosymbionts most commonly found in pea aphids, Hamiltonella defensa, Regiella insecticola, Serratia symbiotica, X-type, Rickettsia, Spiroplasma and Rickettsiella (Oliver et al. 2010; Tsuchida et al. 2010), with polymerase chain reaction (PCR) using symbiont-specific diagnostic primers for the 16S rRNA gene (Table S1). The identity of any symbionts was confirmed by sequencing the PCR product using Big Dye Terminator v.3.1 (Applied Biosystems). In cases of single infections, the PCR product for sequencing was generated using the universal primers 10F and 35R (Table S1), and sequenced using 10F, 1507R and symbiont-specific diagnostic reverse primers (Table S1). In cases of multiple infections, the PCR product was generated using 10F and the appropriate diagnostic reverse primer and sequenced using these two primers (Table S1). Assembled and manually edited sequences for each of the distinct symbiont genotypes were deposited in Genbank (accession nos. JX533642–JX533651). All samples that tested positive for Hamiltonella were also screened for the bacteriophage APSE by amplifying and sequencing five structural genes of the phage and four loci flanking its integration sites into the bacterial genome (Degnan and Moran 2008a, b).
Development of experimental lines
Four Hamiltonella strains from different grain aphid genotypes were used in our experiments; they were named after the original hosts (Table 1). The four strains represented three distinct 16S rRNA genotypes, strains Ha-Co23 and Ha-Co37 being identical at 16S (Table S2). We first removed the experimental symbiont strains from the aphid genotype that they naturally infect by using antibiotics (Łukasik et al. 2011). Each symbiont strain was then reintroduced through microinjection (Chen and Purcell 1997) into its original host genotype, and also introduced into one to three other grain aphid genotypes (as detailed in Table 1) which did not naturally contain any symbionts. Comparisons could then be made between the lines of an aphid genotype that carried no symbionts (naturally or through curing) and lines of the same aphid genotype into which symbionts had been introduced.
To initiate a new line, aphids were microinjected with infected haemolymph and those that survived maintained in the laboratory. Offspring produced during the first 14 days were discarded while subsequent offspring were retained and allowed to reproduce. Aphids from this generation were kept separately and after they had produced offspring they were killed and tested for the presence of the introduced bacteria using diagnostic PCR. Only the offspring of aphids that tested positive were used to start a new line. The presence of the symbiont was confirmed again after eight generations and before the start of the experiments. As a further check against contamination, the identity of the aphid genotype was verified using a panel of microsatellites.
The effects of symbionts on aphid fecundity
Two fecundity experiments were conducted. In the first we tested whether the presence of each of the four Hamiltonella strains affected aphid fecundity in one proven permissive genotype (the one from which the strain originated) and in one of the naturally uninfected genotypes (see Table 1 for details). The experiment was conducted separately for each set of four lines, two infected with a particular symbiont strain and two corresponding uninfected lines. In the second fecundity experiment, we explored the effects of different symbionts in the same aphid genetic background. For two aphid genotypes, neither naturally carrying symbionts, we compared the fecundity of uninfected aphids with three lines of aphids each carrying a different Hamiltonella strain (details in Table 1). All experiments on the same aphid genotype were carried out at the same time.
The fecundity assays followed the protocol described by Łukasik et al. (2011). In each block, aphids were born within the same 6-h period and were transferred individually into Petri dishes with single Triticum seedlings. Every 3 days these aphids were transferred to a fresh plant in a new dish, and any offspring counted. The number of offspring produced in the first 16 days of life was used as a measure of fecundity. Aphids that died during the experiments were excluded from the analysis (there was no evidence that symbiont infection status affected survival). Because the experiment was started before it could be determined whether the aphid was going to be winged or wingless, both morphs were tested. It is well known that this phenotype affects aphid fecundity and we thus only included replicates of aphids of the most common morph in any particular temporal block in the analysis: winged morphs were used for aphid genotypes Co31 and Co37 in experiment 1, and for genotype Co50 in experiment 2; all other aphids were wingless. This limited our ability to compare across symbiont strains (experiment 1) or aphid genotypes (experiment 2) but increased the power to detect the effects of the symbionts on fecundity, the main purpose of the experiments. In experiment 1, we used 8–16 (mean 11.6) replicates per line in the analysis and in experiment 2, we used 13–31 (mean 23.9) replicates per line.
Aphid susceptibility to parasitoids
We measured the effects of Hamiltonella on grain aphid susceptibility to parasitoids in three separate experiments. In the first we worked with four different aphid genotypes that did not originally carry symbionts and compared their susceptibility to parasitoid attack when they were or were not infected with one of the four Hamiltonella strains. Aphids were challenged by two species of aphidiine parasitoids, A. ervi and E. plagiator. The second experiment was designed after we observed little protective effect of Hamiltonella. It used the same aphid genotypes as in experiment 1, but in addition included the aphid genotypes from which the different strains were collected (in their cured and re-injected forms). In this experiment only the parasitoid A. ervi was used. The final experiment was also prompted by the lack of a protective effect of Hamiltonella in S. avenae. The pea aphid genotype N341 harbors Hamiltonella, X-type and Rickettsiella symbionts and is known to have symbiont-mediated parasitoid protection (J. Ferrari, unpublished data). Haemolymph from this genotype was injected into a line of the S. avenae genotype Co26 that had been cured of its original Hamiltonella infection. A stable infection containing Hamiltonella and X-type from the pea aphid donor was established. We then compared the susceptibility to A. ervi of the double-infected and uninfected lines of this genotype of grain aphid. A summary of the genotypes and symbiont strains used in the experiments is provided in Table 1.
A. ervi was obtained from Syngenta Bioline in 2007. A stock culture of E. plagiator was established in September 2008 from parasitoids emerging from mummies of unidentified aphids collected from Holcus sp. near Lower Radley. The identity of both parasitoid species was confirmed by sequencing a fragment of COI gene (Traugott et al. 2008). Both species had been in culture on the naturally symbiont-free S. avenae genotype Co50, maintained on young potted Triticum plants enclosed in 30 × 30 × 30-cm transparent cages, for at least 15 generations before they were used in the experiments.
To assess susceptibility, groups of thirty 72- to 96-h-old aphids (late 2nd or early 3rd instar) were transferred to 90-mm Petri dishes with two Triticum seedlings whose stems were inserted into agar. After the aphids settled on the plants, single mated parasitoid females with no experience of oviposition were introduced into each dish. Aphids were exposed to parasitoids for 8 h in the case of A. ervi and 10 h for E. plagiator. Pilot experiments showed that in these times the wasps were able to parasitize approximately 90 % of the available aphids. In the first experiment replicates in which no parasitism occurred were removed from the analysis (excluded dishes were distributed randomly across aphid genotypes and lines). In the second and third experiments, any parasitoids that did not start stinging aphids within 15 min of introduction were replaced with fresh wasps. After the parasitoids were removed from the dishes, the aphids were kept for 15 days at 20 ± 2 °C. Every 3 days they were moved to fresh dishes, and during each transfer any parasitoid mummies, as well as reproducing and dead aphids, were counted. The proportion of aphids exposed to wasps in which the parasitoids successfully pupated was used as the measure of susceptibility.
In the first experiment, replicates involving each of the two parasitoid species were carried out in two temporal blocks. Parasitoid emergence rate, sex ratio and development times were also measured for each species in one of the two blocks (details in the Electronic Supplementary Material). In the second experiment, replicates involving each symbiont strain were carried out at the same time. There was no temporal blocking in the third experiment. The number of replicates per line in the first susceptibility experiment was from seven to 13 (average 10.9) for A. ervi and from seven to nine (average 7.9) for E. plagiator. In the second susceptibility experiment from five to eight (average 6.1) replicates per line was used and in the third there were from eight to ten (average 9.0) replicates per line.
Parasitoid oviposition choice
We presented experienced females of the parasitoids A. ervi and E. plagiator with the choice of genetically identical aphids carrying and not carrying Hamiltonella and observed the number of eggs oviposited in each type of host. Experiments were carried out with all four Hamiltonella strains in aphid genotypes that did not naturally carry symbionts (see Table 1 for details).
Parasitoids were allowed to obtain experience of oviposition by being placed, soon after emergence, in groups of four females and four males in 140-mm non-vented Petri dishes with Triticum seedlings whose stems were inserted into agar, each containing approximately 250 symbiont-infected and 250 uninfected aphids of all instars. The aphids were of the same genotype as the parasitoids would later encounter in the experiment and the wasps were allowed to search and oviposit for 24 h. The wasps used in the experiment would thus have encountered both uninfected and infected aphids and would also have partly depleted their reserves of mature eggs, which we reasoned would make them more selective.
The choice experiment was conducted in 90-mm Petri dishes containing two young Triticum seedlings with their stems inserted in agar. The aphids to be used in each experiment were reared at low density in multiple, randomly distributed Petri dishes. Infected and non-infected aphids were distinguished by cutting off part of the last segment of either the right or left antenna at least 6 h before the experiment; 15 uninfected and 15 Hamiltonella-infected aphids were used in each replicate. Single parasitoid females were introduced to the dishes once the aphids, which were then 60- to 72-h-old, had settled on the plants. Aphids were exposed to A. ervi for 9 h and to E. plagiator for 12 h. Subsequently, the dishes were frozen and kept at −26 °C until the aphids were dissected and the parasitoid eggs in each aphid counted (Oliver et al. 2003). All replicates with the same parasitoid species were carried out at the same time. Aphids from 17 dishes exposed to A. ervi and 16 dishes exposed to E. plagiator contained parasitoid eggs while no eggs were observed in four dishes which were excluded from the analysis. In order to test whether differences in size between infected and uninfected aphids influenced parasitoid oviposition decisions, the length of a subsample of 2nd-instar aphids exposed to A. ervi (30 aphids per line) was measured under a binocular microscope before dissection.
To study the behavioural mechanisms underlying parasitoid discrimination we observed naïve female A. ervi that were presented with a choice of genetically identical aphids, some carrying and some not carrying Hamiltonella. Full details of the experimental methods are given in the Electronic Supplementary Material.
Statistical analyses
All analyses were conducted using generalised linear modelling techniques implemented in the statistical package R version 2.13.0 (R Development Core Team 2011). For the analysis of fecundity data Gaussian error variance was assumed after the data distribution was checked for normality. Other count and proportion data were analysed assuming quasi-Poisson and quasi-binomial error variances which account for overdispersion. Parasitoid development times and dry weights were log-transformed prior to analysis assuming Gaussian error variance.
Results
Symbionts in a grain aphid population
Fifty S. avenae were collected at two field sites and genotyped at seven microsatellite loci. The aphids were assigned to 22 distinct genetic lineages (henceforth referred to as “genotypes”) (Table S2). Eight of these genotypes were represented by multiple individuals and six of them were collected from more than one plant species. The seven aphids collected at Lower Radley all belonged to genotypes represented at our main collection site, Great Coxwell.
Thirteen aphids (26 %) carried no facultative endosymbionts while 23 (46 %) were infected with Hamiltonella defensa, 17 (34 %) with Regiella insecticola, and three (6 %) with Serratia species. These numbers included two aphids that hosted two symbiont species each as well as two aphids hosting three symbionts each (Table S2). No cases of infection by X-type, Rickettsia, Spiroplasma or Rickettsiella were observed. Partial sequencing of the 16S rRNA gene revealed three distinct genotypes of Regiella and four genotypes of Hamiltonella, all at least 99 % identical to the reference sequences of the two symbiont species (Moran et al. 2005b). All field-caught aphids harbouring Hamiltonella tested positive for the bacteriophage APSE. Sequences of Serratia in two of the samples were identical to each other and 99.7 % identical to the reference sequence of S. symbiotica (Moran et al. 2005b). The third sequence was 4.3 % divergent and identical to the sequence of Serratia proteamaculans, a bacterium found in a range of habitats including arthropod guts (Kwak et al. 2006), suggesting that it may have belonged to a transient gut bacterium rather than to a facultative endosymbiont. In five out of the eight genotypes collected more than once, field-caught individuals differed in the presence or species of symbionts they hosted (Table S2).
Laboratory cultures were established using 32 lines originating from different field-collected females, together representing 18 aphid genotypes. Most of these cultures were discarded between 4 months and 2 years after collection, but 14 of these lines (one per genotype) were retained in culture for 34 months. Periodic retesting showed natural single infections to be stable, with no spontaneous symbiont losses observed. Similarly, no symbiont losses were observed over 2 years in experimentally established single infections. However, in the originally triple-infected line of genotype Co39 and in the line of genotype Co21 double-infected with Hamiltonella and Regiella (Table S2), only a single symbiont (Hamiltonella in the former case, Regiella in the latter) could be detected after 1 year in culture.
The effects of symbionts on aphid fecundity
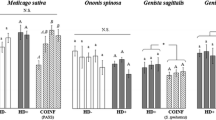
In the first fecundity experiment we asked whether four strains of Hamiltonella influenced the fecundity of: (1) the genotypes from which they had been collected in the field, and (2) naturally symbiont-free genotypes (Table 2). We controlled first for a block effect, inflated because fecundity of either winged or wingless aphids was scored in different blocks; when winged and wingless aphids are analysed separately, the block effect disappears while our other conclusions are unaffected (Table S3). We found that the four originally Hamiltonella-infected genotypes had on average higher fecundity than the four originally symbiont-free genotypes, irrespective of whether they carried the symbiont at the time of the experiment (Table 2; Fig. 1a). Adding present infection status to the statistical model did not significantly improve its fit: there was no evidence of a systematic effect of Hamiltonella on fecundity. However, symbiont strains differed significantly in the effect on fecundity. There was also a significant difference in how the fecundity of different aphid genotypes changed in response to the introduction of the symbiont (a three-way interaction), but this was not due to the aphid’s original infection status (Table 2).
a The effects of infection with one of four strains of Hamiltonella on the fecundity (mean ± SE) of different grain aphid genotypes (fecundity experiment 1). Each symbiont strain, represented by ovals of the same shape and colour, was tested in its original host genotype (white bars) and in a novel, naturally symbiont-free genotype (grey bars). Data are shown for winged (genotypes Co37 and Co31) or wingless (all other genotypes) aphids. Significant differences between infected and uninfected lines of the same genotype, according to a post hoc honestly significant difference (HSD) test, are indicated by asterisks: **P < 0.01, ***P < 0.001. b The effects of three symbiont strains, represented by the same symbols as above, on the fecundity of two originally symbiont-free aphid genotypes (fecundity experiment 2). Data are shown for winged (genotype Co50) or wingless (Co28) aphids. The same letters identify estimates that are not significantly different from others using Tukey’s HSD test with 95 % confidence intervals
In the second fecundity experiment we explored the effects of three symbionts in two originally uninfected aphid genotypes to test whether there was an aphid genotype × bacterial genotype interaction (Fig. 1b). We found no overall effect of hosting Hamiltonella (F 1,185 = 0.90, P = 0.34). There were, however, significant differences between lines infected with different Hamiltonella strains (F 2,183 = 8.01, P < 0.001). In both genotypes, the Hamiltonella strain originating from Co26 had the most negative effect on host fecundity, and that from Co23 the most positive. There were no significant interactions between aphid genotype and infection status (F 1,182 = 3.35, P = 0.07), nor between aphid genotype and specific symbiont strains (F 2,180 = 0.33, P = 0.72).
Aphid susceptibility to parasitoids
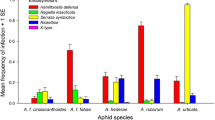
In the first susceptibility experiment, we measured the effects of Hamiltonella infection in four aphid genotypes on the development of two parasitoid species. There was no effect of Hamiltonella on the proportion of aphids succumbing to A. ervi parasitism (F 1,79 = 0.15, P = 0.70) and no genotype × infection status interaction (F 3,78 = 0.94, P = 0.42; Fig. 2). The presence of Hamiltonella had no overall effect on the proportion of aphids successfully parasitized by E. plagiator (F 1,54 = 0.01, P = 0.91), though in this case there were differences amongst aphid genotypes (genotype × infection status interaction: F 3,53 = 5.55, P = 0.002; Fig. 2). Comparisons of parasitoid emergence rates, development times and sizes across aphid lines showed no consistent effects of symbionts (see Electronic Supplementary Material for details).
The effect of Hamiltonella on grain aphid susceptibility to two species of parasitoids. The rate (mean ± SE) of successful parasitism of four grain aphid genotypes, either naturally free of infection with secondary symbionts or artificially infected with different strains of Hamiltonella, is shown. Different symbiont strains are represented by ovals of the same shape and colour
In the second susceptibility experiment, the effect of Hamiltonella on resistance to A. ervi in genotypes that were or were not naturally infected was studied. Again, we found no protective effect of the symbiont (F 1,90 = 1.00, P = 0.32). No differences were found in susceptibility between originally infected and uninfected genotypes, nor between symbiont strains (P > 0.10; Fig. S1).
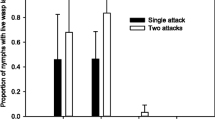
To ascertain whether parasitoid protection by facultative endosymbionts is possible in S. avenae we introduced symbionts (Hamiltonella and X-type) known to confer parasitoid protection in the pea aphid into grain aphid genotype Co26. These symbionts reduced the rate of successful pupation of A. ervi from 64 to 0 % (F 1,17 = 252.3, P < 0.001; Fig. 3).
Parasitoid oviposition choice
We tested whether experienced females of the two parasitoid species preferentially oviposit in symbiont-free aphids. We detected a significantly negative effect of symbiont infection on the probability of parasitism by both A. ervi (χ2 = 8.52, df = 1, P = 0.004) and E. plagiator (χ2 = 7.87, df = 1, P = 0.005), with Hamiltonella-infected aphids having on average a 12.8 % greater chance of avoiding parasitism by the former and 11.4 % by the latter parasitoid. The strength of parasitoid choice was significantly influenced by aphid genotype in E. plagiator (χ2 = 8.83, df = 3, P = 0.032), and there was a non-significant tendency for this to occur in A. ervi (χ2 = 6.22, df = 3, P = 0.101) (Fig. 4).
Oviposition preferences of two parasitoid species when offered the choice of Hamiltonella-infected and symbiont-free aphids. The graph shows the proportion (mean ± SE) of grain aphids that were found to contain at least one parasitoid egg after mixed groups of infected and uninfected aphids were exposed to single parasitoid females. Data are shown for all experimental genotypes combined (diagonally striped bars), as well as separately for each of the aphid genotypes. Infected lines are marked with ovals. Significant differences between infected and uninfected aphids within a comparison are indicated by asterisks: **P < 0.01, ***P < 0.001
The mean proportion of aphids that contained at least one parasitoid egg was 0.46 among aphids exposed to A. ervi, and 0.64 among aphids exposed to E. plagiator. In 91 % of cases, E. plagiator females laid only a single egg in each aphid they parasitized, and we never found more than two eggs per aphid. A. ervi superparasitized 41.5 % of experimental S. avenae, with up to six eggs in a single aphid. The mean number of A. ervi eggs in parasitized aphids was not significantly affected by the presence of the symbiont, and there were no differences between aphid genotypes in how symbiont infection affected the number of eggs (P > 0.15). In the eight A. ervi exposure dishes for which we collected data on the size of the exposed aphids, parasitoid females tended to oviposit more eggs into larger aphids (F 4,211 = 2.08, P = 0.084). However, there were no differences in the average sizes of the infected and uninfected aphids from the same dishes, and no differences between genotypes in the effects of Hamiltonella on aphid size (P > 0.50).
Direct observations of the behaviour of naïve parasitoids that were offered infected and uninfected aphids simultaneously did not indicate parasitoid bias or different behaviour towards either type of aphid, nor differences in the defensive behaviour of the aphids (see Electronic Supplementary Material for details).
Discussion
We found that the grain aphid S. avenae hosts at least three of the seven facultative endosymbionts frequently found in the much more extensively studied pea aphid. The most common symbiont was Hamiltonella, but in marked contrast to the pea aphid, it did not confer physiological resistance against parasitoids. Despite this, parasitoid females preferentially oviposited into uninfected aphids. We found no systematic effect of infection on S. avenae fecundity.
The symbionts which we detected in grain aphids included strains of three of the seven species known from the pea aphid: Hamiltonella defensa, Regiella insecticola and Serratia symbiotica (Moran et al. 2005b). Two of them, Hamiltonella and Regiella, have recently been reported from S. avenae collected in Germany (Alkhedir et al. 2013). In 43 grain aphids collected at our main sampling site, we detected four distinct 16S genotypes of Hamiltonella and three of Regiella (Table S2). This genetic diversity appears to be of the same order as that found in a survey of 297 pea aphid genotypes representing eight genetically distinct host plant races from England and Germany (Ferrari et al. 2012). Looking at genes less conserved than 16S could well reveal even higher symbiont strain diversity within and across aphid populations (Russell et al. 2013). We found that the same grain aphid genotypes frequently hosted different symbiont types. This, as well as our observations of symbiont transmission failure in natural multiple infections, provides an insight into the dynamics of aphid-symbiont associations in the field and suggests that transmission failure and possibly even horizontal transmission can occur on ecological timescales (Leonardo 2004; Oliver et al. 2010).
Hamiltonella, the most common of the grain aphid facultative endosymbionts, has been shown to confer protection against hymenopterous parasitoids in pea, black bean and cowpea aphids (Desneux et al. 2009; Oliver et al. 2003, 2005; Vorburger et al. 2009). We hypothesised that S. avenae, a species regularly attacked by hymenopterous parasitoids (Schmidt et al. 2003; Sigsgaard 2002), was also likely to enjoy protection due to Hamiltonella from these natural enemies. However, none of the four Hamiltonella strains conferred resistance against either of the parasitoid species in our experiments (Figs. 2, S1). Furthermore, symbiont infection had no consistent effects on parasitoid development time or adult size (Fig. S2), the two traits which had been shown to be negatively affected in wasps emerging from resistant pea aphids and black bean aphids (Li et al. 2002; Nyabuga et al. 2010; Schmid et al. 2012). However, a Hamiltonella strain originating from the pea aphid did confer resistance in S. avenae (though in the presence of an additional symbiont, X-type). This suggests that there are functional differences between the Hamiltonella strains from S. avenae and A. pisum, even though they are more than 99 % identical at the 16S rRNA gene. Strains of Hamiltonella from pea aphid that do not confer resistance are known, and this seems to be associated with the absence of the APSE bacteriophage (Degnan and Moran 2008b; Moran et al. 2005a; Oliver et al. 2009). However, all the Hamiltonella strains isolated from S. avenae carried APSE. S. avenae is attacked by at least 17 species of hymenopterous primary parasitoids whose abundances fluctuate widely across seasons, years and geographic areas (e.g. Feng et al. 1991; Kavallieratos et al. 2004; Muller et al. 1999; Powell 1982). It is possible that the four Hamiltonella strains that we studied protect their hosts from some of these other species. They could also confer protection against the two experimental parasitoids but under different conditions from those used here, for example at lower temperatures (Bensadia et al. 2006). Nevertheless, our observations that none of the four experimental strains of Hamiltonella had an effect on fitness of the two parasitoids of S. avenae, one of which was common in the aphid communities that we sampled in 2008 (E. plagiator; P. Łukasik, unpublished data), shows that this symbiont does not universally protect its host against hymenopterous parasitoids. It will be interesting to further investigate the genetic and functional basis of the differences in defensive properties of symbionts in a range of aphid species.
Even though aphid symbionts appear not to harm parasitoids in S. avenae, experienced females of both wasp species preferentially oviposited into Hamiltonella-free aphids when given a choice between infected and uninfected insects. Parasitoids may be selected to avoid symbiont-infected aphids if their lack of a protective effect in Sitobion is unusual. Aphid parasitoids are known to discriminate between prospective hosts from different species (Daza-Bustamante et al. 2003; Henry et al. 2008), of different sizes (Henry et al. 2006, 2009), and based on whether the hosts have previously been parasitised by the same or a different species (Outreman et al. 2001). Recently, Oliver et al. (2012) demonstrated that A. ervi females discriminate between pea aphids based on the symbionts that they carry. They found that wasps preferentially superparasitized hosts carrying Hamiltonella and that this increased the probability of successful parasitism. One explanation for the seemingly unnecessary avoidance behaviour of A. ervi and E. plagiator in our experiments is that both species are relatively polyphagous and likely parasitize other aphid species where Hamiltonella may increase resistance (Daza-Bustamante et al. 2003; Kavallieratos et al. 2004). The different responses of parasitoids to Hamiltonella infection in our and Oliver et al.’s (2012) experiments—avoidance versus superparasitism—can be interpreted as instances of parasitoids identifying infected aphids as inferior hosts, and adjusting their oviposition decisions accordingly. We used experienced parasitoids in our experiments while Oliver et al. (2012) worked with naïve females. Our wasps would already have begun to deplete their egg reserves and had experienced an environment rich in high-quality hosts, while the naïve wasps would be carrying more eggs, and due to not having encountered hosts before may be less able to predict the quality of environment. Thus our wasps may have responded to the discovery of inferior hosts by avoiding oviposition and conserving egg reserves while those in Oliver et al.’s (2012) experiment may have “invested” excess eggs in overcoming host defences.
While our experiments found no evidence for a role of Hamiltonella in host protection, it may affect fitness in other ways that could explain its maintenance in S. avenae populations. However, no such clear effects have been demonstrated to date (Alkhedir et al. 2013; Łukasik et al. 2011); in particular, we detected no consistent effects of Hamiltonella on grain aphid fecundity. In the analysis of fecundity we did find a significant interaction between host genotype × symbiont presence (Fig. 1) and similar complex genetic interactions have been observed in the analysis of the effects of symbionts on other components of fitness in pea aphids and black bean aphids (Chen et al. 2000; McLean et al. 2011; Oliver et al. 2008; Vorburger and Gouskov 2011). Curiously, the grain aphid genotypes we collected that originally carried Hamiltonella had higher fecundity than those that were originally symbiont-free, irrespective of whether they carried the bacterium at the time of testing. Similar observations have been made in the black bean aphid (Castaneda et al. 2010; Vorburger et al. 2009). A possible explanation could be that the aphid genotypes which perform better on experimental plant varieties belong to host plant-adapted races. In the pea aphids such races tend to be associated with particular symbiont types, which however do not consistently influence host fecundity under permissive laboratory conditions (Ferrari et al. 2012; McLean et al. 2011).
It is becoming clear that facultative endosymbiotic bacteria have major effects on the ecology and evolution of their arthropod hosts, and can influence many aspects of their biology. Work on the pea aphid and its lengthening list of bacterial associates revealed the role of facultative endosymbiotic bacteria protecting their hosts from natural enemies (Brownlie and Johnson 2009; Haine 2008; Łukasik et al. 2013; Oliver et al. 2010). The results of the present study indicate that there may be considerable differences between related species in the extent to which they rely on symbiotic bacteria for protection. This could have significant effects not only on the biology of individual species, but also on their polyphagous natural enemies and on indirect interactions between herbivores mediated by shared pathogens and parasitoids. In order to comprehend fully the evolutionary and ecological processes shaping arthropod communities, it is thus essential to explore the roles of symbiotic bacteria beyond the well-characterized model systems.
References
Alkhedir H, Karlovsky P, Vidal S (2013) Relationship between water soluble carbohydrate content, aphid endosymbionts and clonal performance of Sitobion avenae on cocksfoot cultivars. PLoS One 8:e54327
Bensadia F, Boudreault S, Guay J-F, Michaud D, Cloutier C (2006) Aphid clonal resistance to a parasitoid fails under heat stress. J Insect Physiol 52:146–157
Brownlie JC, Johnson KN (2009) Symbiont-mediated protection in insect hosts. Trends Microbiol 17:348–354
Burke GR, Normark BB, Favret C, Moran NA (2009) Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol 75:5328–5335
Castaneda LE, Sandrock C, Vorburger C (2010) Variation and covariation of life history traits in aphids are related to infection with the facultative bacterial endosymbiont Hamiltonella defensa. Biol J Linn Soc 100:237–247
Chen DQ, Montllor CB, Purcell AH (2000) Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl 95:315–323
Chen DQ, Purcell AH (1997) Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol 34:220–225
Daza-Bustamante P, Fuentes-Contreras E, Niemeyer HM (2003) Acceptance and suitability of Acyrthosiphon pisum and Sitobion avenae as hosts of the aphid parasitoid Aphidius ervi (Hymenoptera : Braconidae). Eur J Entomol 100:49–53
Dedryver C-A, Le Ralec A, Fabre F (2010) The conflicting relationships between aphids and men: a review of aphid damage and control strategies. Comptes Rendus Biol 333:539–553
Degnan PH, Moran NA (2008a) Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol 74:6782–6791
Degnan PH, Moran NA (2008b) Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol Ecol 17:916–929
Desneux N, Barta RJ, Hoelmer KA, Hopper KR, Heimpel GE (2009) Multifaceted determinants of host specificity in an aphid parasitoid. Oecologia 160:387–398
Dion E, Zele F, Simon JC, Outreman Y (2011) Rapid evolution of parasitoids when faced with the symbiont-mediated resistance of their hosts. J Evol Biol 24:741–750
Duron O et al (2008) The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol 6:12
Engelstadter J, Hurst GDD (2009) The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst 40:127–149
Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543
Feng MG, Johnson JB, Halbert SE (1991) Natural control of cereal aphids (Homoptera, Aphididae) by entomopathogenic fungi (Zygomycetes, Entomophthorales) and parasitoids (Hymenoptera, Braconidae and Encyrtidae) on irrigated spring wheat in southwestern Idaho. Environ Entomol 20:1699–1710
Ferrari J, Vavre F (2011) Bacterial symbionts in insects or the story of communities affecting communities. Philos Trans R Soc B Biol Sci 366:1389–1400
Ferrari J, West JA, Via S, Godfray HCJ (2012) Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66:375–390
Foottit RG, Maw HEL, Von Dohlen CD, Hebert PDN (2008) Species identification of aphids (Insecta: Hemiptera: Aphididae) through DNA barcodes. Mol Ecol Resour 8:1189–1201
Fukatsu T, Tsuchida T, Nikoh N, Koga R (2001) Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta : Homoptera). Appl Environ Microbiol 67:1284–1291
Guay JF, Boudreault S, Michaud D, Cloutier C (2009) Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol 55:919–926
Haine ER (2008) Symbiont-mediated protection. Proc R Soc B Biol Sci 275:353–361
Haynes S, et al. (2003) Diversity of bacteria associated with natural aphid populations. Appl Environ Microbiol 69:7216–7223
Henry LM, Ma BO, Roitberg BD (2009) Size-mediated adaptive foraging: a host-selection strategy for insect parasitoids. Oecologia 161:433–445
Henry LM, Roitberg BD, Gillespie DR (2006) Covariance of phenotypically plastic traits induces an adaptive shift in host selection behaviour. Proc R Soc B Biol Sci 273:2893–2899
Henry LM, Roitberg BD, Gillespie DR (2008) Host-range evolution in Aphidius parasitoids: fidelity, virulence and fitness trade-offs on an ancestral host. Evolution 62:689–699
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol Lett 281:215–220
Himler AG et al (2011) Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332:254–256
Kavallieratos NG et al (2004) A survey of aphid parasitoids (Hymenoptera: Braconidae: Aphidiinae) of Southeastern Europe and their aphid-plant associations. Appl Entomol Zool 39:527–563
Kwak J et al (2006) Polyphasic assignment of a highly proteolytic bacterium isolated from a spider to Serratia proteamaculans. J Microbiol Biotechnol 16:1537–1543
Leonardo TE (2004) Removal of a specialization-associated symbiont does not affect aphid fitness. Ecol Lett 7:461–468
Leonardo TE, Mondor EB (2006) Symbiont modifies host life-history traits that affect gene flow. Proc R Soc B Biol Sci 273:1079–1084
Li S et al (2002) Pea aphid clonal resistance to the endophagous parasitoid Aphidius ervi. J Insect Physiol 48:971–980
Łukasik P, Hancock EL, Ferrari J, Godfray HCJ (2011) Grain aphid clones vary in frost resistance, but this trait is not influenced by facultative endosymbionts. Ecol Entomol 36:790–793
Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ (2013) Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecology Lett 16:214–218
McLean AHC, van Asch M, Ferrari J, Godfray HCJ (2011) Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc R Soc B Biol Sci 278:760–766
Montllor CB, Maxmen A, Purcell AH (2002) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195
Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H (2005a) The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci USA 102:16919–16926
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190
Moran NA, Russell JA, Koga R, Fukatsu T (2005b) Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol 71:3302–3310
Muller CB, Adriaanse ICT, Belshaw R, Godfray HCJ (1999) The structure of an aphid-parasitoid community. J Anim Ecol 68:346–370
Nyabuga FN, Outreman Y, Simon JC, Heckel DG, Weisser WW (2010) Effects of pea aphid secondary endosymbionts on aphid resistance and development of the aphid parasitoid Aphidius ervi: a correlative study. Entomol Exp Appl 136:243–253
Oliver KM, Campos J, Moran NA, Hunter MS (2008) Population dynamics of defensive symbionts in aphids. Proc R Soc B Biol Sci 275:293–299
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266
Oliver KM, Degnan PH, Hunter MS, Moran NA (2009) Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325:992–994
Oliver KM, Moran NA, Hunter MS (2005) Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci USA 102:12795–12800
Oliver KM, Noge K, Huang EM, Campos JM, Becerra JX, Hunter MS (2012) Parasitic wasp responses to symbiont-based defense in aphids. BMC Biol 10:11
Oliver KM, Russell JA, Moran NA, Hunter MS (2003) Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA 100:1803–1807
Outreman Y, Le Ralec A, Plantegenest M, Chaubet B, Pierre JS (2001) Superparasitism limitation in an aphid parasitoid: cornicle secretion avoidance and host discrimination ability. J Insect Physiol 47:339–348
Powell W (1982) The identification of hymenopterous parasitoids attacking cereal aphids in Britain. Syst Entomol 7:465–473
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Russell JA, Latorre A, Sabater-Muńoz B, Moya A, Moran NA (2003) Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol 12:1061–1075
Russell JA, Moran NA (2006) Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc B Biol Sci 273:603–610
Russell JA et al (2013) Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol Ecol (online early)
Sandstrom JP, Russell JA, White JP, Moran NA (2001) Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228
Scarborough CL, Ferrari J, Godfray HCJ (2005) Aphid protected from pathogen by endosymbiont. Science 310:1781
Schmid M, Sieber R, Zimmermann Y-S, Vorburger C (2012) Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct Ecol 26:207–215
Schmidt MH, Lauer A, Purtauf T, Thies C, Schaefer M, Tscharntke T (2003) Relative importance of predators and parasitoids for cereal aphid control. Proc R Soc Lond Ser B Biol Sci 270:1905–1909
Sigsgaard L (2002) A survey of aphids and aphid parasitoids in cereal fields in Denmark, and the parasitoids’ role in biological control. J Appl Entomol Zeitschr Angew Entomol 126:101–107
Traugott M et al (2008) Endoparasitism in cereal aphids: molecular analysis of a whole parasitoid community. Mol Ecol 17:3928–3938
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989
Tsuchida T et al (2010) Symbiotic bacterium modifies aphid body color. Science 330:1102–1104
Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T (2002) Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol 11:2123–2135
Vorburger C, Gouskov A (2011) Only helpful when required: a longevity cost of harbouring defensive symbionts. J Evol Biol 24:1611–1617
Vorburger C, Sandrock C, Gouskov A, Castañeda LE, Ferrari J (2009) Genotypic variation and the role of defensive endosymbionts in an all-parthenogenetic host–parasitoid interaction. Evolution 63:1439–1450
Acknowledgments
P. Ł., J. F., H. C. J. G. designed the experiments. P. Ł., M. A. D. performed the experiments. P. Ł., J. F., H. C. J. G. analyzed the data and wrote the manuscript. We thank E. Frago, K. Oliver, J. Russell and K. Sullam for discussion and helpful comments on the manuscript. The work was supported by the UK Biotechnology and Biology Research Council award BB/E010857/1 and by a Sir Richard Southwood (Christensen Foundation) Graduate Scholarship to P. Ł.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by George Heimpel.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Łukasik, P., Dawid, M.A., Ferrari, J. et al. The diversity and fitness effects of infection with facultative endosymbionts in the grain aphid, Sitobion avenae . Oecologia 173, 985–996 (2013). https://doi.org/10.1007/s00442-013-2660-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-013-2660-5