Abstract
The dynactin p150Glued subunit, encoded by the gene DCTN1, is part of the dynein-dynactin motor protein complex responsible for retrograde axonal transport in motor neurons. The p150 subunit is a candidate gene for neurodegenerative diseases, in particular motor neuron and extrapyramidal diseases. Tubulin-binding cofactors are believed to be involved in tubulin biogenesis and degradation and therefore to contribute to microtubule functional diversity and regulation. A yeast-two-hybrid screen for putative interacting proteins of dynactin p150Glued has revealed tubulin-folding cofactor B (TBCB). We analyzed the interaction of these proteins and investigated the impact of this complex on the microtubule network in cell lines and primary hippocampal neurons in vitro. We especially concentrated on neuronal morphology and synaptogenesis. Overexpression of both proteins or depletion of TBCB alone does not alter the microtubule network and/or neuronal morphology. The demonstration of the interaction of the transport molecule dynactin and the tubulin-regulating factor TBCB is thought to have an impact on several cellular mechanisms. TBCB expression levels have been found to have only a subtle influence on the microtubule network and neuronal morphology. However, overexpression of TBCB leads to the decreased localization of p150 to the microtubule network that might result in a functional modulation of this protein complex.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cytoskeleton in eukaryotic cells is a crucial structural framework that serves as a communication and transport module within cells to ensure their survival and growth. In addition to their maintenance of cellular morphology and polarization, cytoskeletal protein complexes are utilized, for example, as migration devices, transport tracks, or classifiers of subcellular compartments. The cytoskeleton is composed of three major kinds of filaments known to date: microfilaments (mainly actin filaments), intermediate filaments, and microtubules. The last-mentioned associate with several proteins, such as the dynactin p150 subunit of the dynactin complex, tubulin-binding cofactor B (TBCB), and many other microtubule-associated proteins, to enable the tubules to execute their functions. TBCB, as its name suggests, is one of a group of the cofactor family (TBCA-TBCE) that plays a role in microtubule biosynthesis. It specifically has been shown to enhance the folding of alpha tubulin (Tian et al. 1997) and also to polymerize microtubules (Vadlamudi et al. 2005). Unlike some members of the tubulin cofactor family that have been implicated in disease via mutational defects, no molecular aberration in TBCB has been discovered so far. Moreover, little is known about the molecular function of this protein.
Dynactin is a macroprotein complex molecule that consists of several protein subunits. Most often, it couples to dynein, a retrograde transport protein. The coupling of dynactin and dynein forms another complex commonly referred to as the dynactin/dynein motor complex, which has been earmarked notably in the transport of molecules along the neuronal axon toward the soma (Burakov and Nadezhdina 2006). The dynactin p150Glued subunit, the largest of the dynactin complex subunits, harbors the dynein motor and microtubule-binding domains to which it binds (Puls et al. 2003). Thus, p150 is thought to serve as a bridge between dynein and microtubules during cargo transportation. In general terms, p150 is involved in the intracellular transport of cargoes (Schroer 2004), such as vesicles, RNA, and trophic factors, that are indispensable for cellular metabolism and survival. However, p150 has been implicated in neurodegeneration (Farrer et al. 2009; Laird et al. 2008; LaMonte et al. 2002) and therefore is considered as a candidate gene in motor neuropathies. The identification of a mutation, G59S, in its conserved cytoskeleton-associated glycine-rich (CAP-Gly) domain, causing motor neuron disease (Levy et al. 2006; Puls et al. 2003), makes this region important for further investigations. Moreover, a common mutant variant of p150, namely the valine substitution of isoleucine at position 196 (I196V), is reported in patients and in healthy controls (Munch et al. 2007); as this mutation shows no clear penetrance, it is therefore considered as only a putative disease-causing factor (Vilarino-Guell et al. 2009).
The direct interaction of p150 and TBCB points to a functional correlation of the tubulin-dependent transport mechanism of the dynein/dynactin complex and the tubulin assembly modulation factor TBCB. In particular, the influence of p150 proteins harboring sequence aberrations on native and artificially overexpressed TBCB is of high importance for the understanding of neuronal degeneration in MND patients carrying DCTN1 mutations. Here we present the direct binding of p150Glued and the tubulin modulator TBCB, providing for the first time, at least to our knowledge, insights into the relationship between these proteins and their impact on cell morphology.
Materials and methods
Ethics statement
All animal experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the Federal Government of Germany, the National Institutes of Health, and the Max Planck Society. The experiments in this study were approved by the review board of the Land Baden Württemberg (permit no. O.103).
Cloning
Truncated human wild-type (WT) p150Glued; exon1-7 was amplified by the polymerase chain reaction and cloned into the yeast vector pGBKT7 (both Clontech). Human WT TBCB and DCTN1 were also amplified from a human fetal male cDNA library and cloned into pEGFP-C3 and PCMV-Myc-C vector systems (both Clontech). Human DCTN1 mutants were generated in a pEGFP-C3 vector. The mouse WT dctn1 gene was amplified from a mouse cDNA library and cloned into the pIRES2 AcGFP-Nuc vector, transcribing a bicistronic mRNA for expressing an unaffected non-fluorescent protein of interest and a green fluorescent protein (GFP) targeted to the nucleus as a transfection control. Mutants were obtained by site-directed mutagenesis.
Gene knockdown
TBCB gene repression targeting sequences were cloned into the pSUPER.retro.neo vector expressing a GFP-tag (OligoEngine, Seattle, Wash., USA) and were transfected into HEK293T cells at various time points. Cell lysates were obtained (see below), and knockdown was investigated by Western blot analysis. Knockdown sequences were:
-
RNAi1:5’GATCCCCCCCTGCTTCAGCTCCTAGCTCTTCAAGAGAGAGCTAGGAGCTGAAGCAGGGTTTTTGGAAA3’
-
RNAi3:5’GATCCCCCCATCGCTGAGTTCAAGTGTATTCAAGAGATACACTTGAACTCAGCGATGGTTTTTGGAAA3’
Cell culture
Preparation of primary hippocampal neurons (pHNs) from rat embryos (E18) was adopted from Liebau et al. (2009) and Proepper et al. (2007). In brief, after preparation, the hippocampal neurons were seeded on poly-L-lysine (0.1 mg/ml; Sigma-Aldrich, Steinheim, Germany)-coated coverslips at a density of 4×104 cells/well (transfection experiments) or 2×104 cells/well (immunocytochemical staining). Cells were grown in Neurobasal medium complemented with B27 supplement, 0.5 mM L-glutamine, and 100 U/ml penicillin/streptomycin (all Invitrogen, Karlsruhe, Germany) and maintained at 37 °C in 5 % CO2. Hippocampal cells were transfected on the days indicated by using Lipofectamine 2000 according to the manufacturer’s recommendation (Invitrogen). Cos7 and HEK293T cells (obtained from DSMZ, Braunschweig, Germany) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose (Invitrogen) supplemented with 10 % (v/v) fetal calf serum and 2 mM L-glutamine without antibiotics. Cells were grown on commercially available chamber-slides (Nunc, Wiesbaden, Germany) treated with poly-L-lysine (0.1 mg/ml; Sigma-Aldrich).
Transfections
COS7 and HEK293T were transfected for immunocytochemistry with Polyfect (Qiagen) according to the manufacturer’s protocol. Hippocampal neurons were transfected with Optifect (Invitrogen) according to Liebau et al. (2009). Respective constructs were overexpressed for 24, 48, 72, and 96 h in culture.
Immunoprecipitation (pull-down)
Cell extracts of 293 T cells cotransfected with human p150 mutants and myc-labeled WT TBCB, prepared with Triton X-100 lysis buffer (150 mM NaCl, 1 % Triton-X100, 50 mM TRIS-HCl pH 8.0, 1× protease inhibitors cocktail) were coupled to μMACS magnetic Microbeads coated with anti-myc antibody (MiltenyiBiotec, Germany) and washed in a magnetic field. The magnetized complex was eluted with sodium dodecyl sulfate (SDS) gel loading buffer heated to 95 °C. Eluates were probed for p150 by using anti-GFP antibody in Western blots. The coupling of lysate with anti-GFP-coated magnetic Microbeads was also performed to confirm the results in both directions. Eluates were used for Western blot.
Western blot
Protein sample concentrations were determined by the amido black protein assay. Samples of 10 μg protein were loaded per lane and separated by standard SDS electrophoresis by using a polyacrylamide gel with 10 % total monomer concentration. Electro-blotting onto polyvinylidene difluoride membranes was conducted by standard protocols. Primary antibodies used were: anti-GFP (1:3000; Clontech), anti-myc (1:3000; Roche, Mannheim, Germany), anti-alpha tubulin (1:5000; Sigma-Aldrich, Steinheim, Germany), anti-acetylated alpha tubulin (1:2500; Sigma-Aldrich), anti-p150 (1:500; BD Transduction, San Jose, USA), anti-TBCB (1:1000; Sigma-Aldrich). Secondary antibodies used were: anti-rabbit immunoglobulins (IgG; 1:2000; Dako, Hamburg, Germany) and anti-mouse IgG (1:8000; Dako), both linked to horseradish peroxidase (HRP). Protein bands were detected by using enhanced chemiluminescent (ECL) substrate for the detection of HRP (Thermo Fisher, Langenselbold, Germany).
Immunocytochemistry
Immunocytochemistry was performed according to Kleger et al. (2010), Liebau et al. (2011), and Linta et al. (2012). In brief, cultured cells were fixed with 4 % paraformaldehyde (PFA)/1.5 % sucrose/phosphate-buffered saline (PBS) for 15 min at room temperature (RT). After being washed three times with 1× PBS for 5 min at RT, the cells were permeabilized for 3 min on ice in a buffer containing 0.2 % Triton X-100/0.1 % Na-citrate/PBS and washed again three times with 1× PBS. Blocking was performed with 10 % fetal calf serum/PBS for 1 h at RT followed by incubation with the primary antibodies, diluted in PBS, for 1 h. After three further washing steps in PBS, the cells were incubated with secondary antibody conjugates for 45 min at RT, washed three times with 1× PBS and then with twice-distilled water for 3 min, and mounted in Vectashield aqueous mount (Vector, Burlingame, USA). Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Primary antibodies against the following antigens were used: dynactin p150 mouse (1:250; BD), dynactin (H-300) rabbit (1:250; Santa Cruz, Santa Cruz, USA), TBCB rabbit (1:500; Sigma-Aldrich), alpha tubulin mouse (1:1000; Sigma-Aldrich), alpha tubulin rabbit (1:1000; Epitomics, Calif., USA), dynactin p50 mouse (1:500; BD), dynactin p62 goat (1:100; Santa Cruz), Arp-1 goat (1:100; Santa Cruz). Fluorescently labeled secondary antibodies were Alexa Fluor 488 (green, with filter set: excitation BP 450–490 nm, FT 510, emission BP 515–565 nm), Alexa Fluor 568 (red, with filter set: excitation BP 534–558 nm, FT 560, emission BP 575–640 nm), and Alexa Fluor 647 (magenta, with filter set: excitation BP 610–670 nm, FT 660, emission BP 640–740 nm; all from Invitrogen and all diluted 1:500). Images were captured by using an upright fluorescence microscope Axioskop2 equipped with a Zeiss charge-coupled device camera (16 bits; 1280×1024 pixels per Image) or the confocal microscope LSM 710 (both Zeiss, Oberkochen, Germany) and analyzed by using Axiovision software (Zeiss).
Yeast-two-hybrid screen
Saccharomyces cerevisiae yeast strains were used, and screening was based on the generation of a functional GAL4 transcription factor that drives the specific lacZ reporter gene. Cloned human p150 WT Exon 1–7 in a pGBKT7 yeast vector were transformed into the AH109 yeast strain, serving as bait. The male fetal human cDNA library, cloned into pACT2 and pretransformed into the Y187yeast strain (Clontech), served as prey. Following overnight cultivations of the bait in SD-Trp at 200 rpm and 30 °C, mating between the two was performed in YPD medium overnight at 40 rpm and 30 °C. Mated cells were harvested at 1000g for 10 min and resuspended in 20 ml YPD. This solution was plated on close to fifty 150-mm selection plates (SD –His, -Leu, -Trp + 3-AT). Colonies positive for interaction were detected by a blue coloration after being subjected to a Filter Lift Assay (FLA). DNA from blue colonies was extracted, and interacting partners were identified by sequencing.
In situ hybridization
Radioactively labeled cDNA probe was added to 60 μl salmon testes DNA and incubated for 5 min at 37 °C followed by 3 min on ice. Subsequently, 600 μl hybridization cocktail and 30 μl tRNA were added to the probe solution; 60 μl of this hybridization solution was pipetted onto each section avoiding bubbles and incubated overnight at 42 °C. After being washed with 1× standard sodium citrate (SSC) for 15 min at RT, sections were rinsed five times for 15 min in wash buffer at 50 °C. Samples were cooled down in wash buffer for 15 min at RT, followed by washing three times with 1× SSC at RT, 2 min each. Dehydration of the sections was performed at RT by transferring the sections to increasing grades of ethanol for 15 s and absolute ethanol for 15 s. The dried sections were then placed into an X-ray film cassette and transferred to a dark room. Exposure of the sections to an X-ray film was allowed to take place for 8–24 days before the film was finally developed.
Statistical analyses
Unless stated otherwise, standard errors of the mean (SEM) are indicated by error bars. Levels of significance were calculated with the unpaired Student t-test or with the analysis of variance model for multiple-group comparisons with post-hoc t-test and Bonferroni adjustment. P-values of <0.05 were considered significant. Calculations of significance were performed with GraphPad Prism 5 (GraphPad Software, La Jolla, Calif., USA) or StatView 5.0 (SAS, Chicago, Ill., USA). All experiments were performed at n=3 as independent experiments and preparations. Colocalization of interacting proteins with different fluorescent signals could be vividly observed. Micrographs were also used for further analysis such as determination of synapse density, dendrite and branching point number in neurons, as performed in Grabrucker et al. (2009) and Liebau et al. (2011). At least 20 cells were chosen randomly for quantification from at least three independent experiments for each condition.
Results
Functional domains and protein and mRNA expression profiles of dynactin p150Glued and TBCB in mammalian tissues
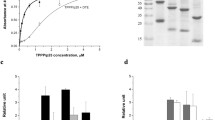
The dynactin subunit (DCTN1) p150 gene product is composed of several domains, some of them known as protein interaction motifs. Noteworthy in this respect are the tubulin-binding motif, the dynein-binding site, the C-terminal cargo-binding motifs, and an N-terminal CAP-Gly domain and two coiled-coil domains. TBCB has a ubiquitin-binding site, a coiled-coil domain, and a C-terminal CAP-Gly domain (Fig. 1a). The endogenous expression of both proteins in the central nervous system (CNS) and throughout the organism is quite identical. Tissue Western blotting showed that both proteins were expressed in all investigated organs with a high expression in the brain (Fig. 1b). CNS protein levels of p150 were similar, whereas protein levels of TBCB varied among the different regions. TBCB showed the highest protein levels in the cortex, and hippocampus (Fig. 1c). Investigations of both proteins in the cellular subcompartments found p150 in all subfractions and even the postsynaptic density (PSD). TBCB was not detected in the synaptic junctions or the PSD (Fig. 1d). mRNA levels for DCTN1 were apparently high throughout all developmental stages in the cortex, cerebellum, dentate gyrus, and hippocampal formation. Whole-mount in situ embryos showed a predominant abundance of DCTN1 in the CNS (Fig. 1e). The distribution of TBCB mRNA in the brain was higher than that of DCTN1 with a higher expression in the medullar compartment. TBCB mRNA was ubiquitously expressed throughout the organs in the whole-mount mRNA in situ specimens (Fig. 1f).
Ubiquitous expression of p150Glued and tubulin-folding cofactor B (TBCB) in mammalian adult tissues and during development. a Representation of p150Glued and TBCB. p150 is five times larger than TBCB and consists of an N-terminal cytoskeleton-associated glycine-rich (CAP-Gly) domain and two coiled-coil domains. TBCB has a functional N-terminal ubiquitin domain, central coiled-coil domain, and a C-terminal CAP-Gly domain. b Western blot detection of endogenous TBCB and p150 from pregnant adult female rat tissues (Br brain, SC spinal cord, Ht heart, Li liver, Sp spleen, Kd kidney, Lu lung, Mu abdominal muscle). The expression of both proteins is ubiquitous with a high expression observed in the brain (GAPDH D-glyceraldehyde-3-phosphate dehydrogenase control). c p150 and TBCB expression in brain subregions. Western blot of homogenates of an adult female rat brain subregions (PFC prefrontal cortex, CTX cortex, HIP hippocampus, STR striatum, THA thalamus, MES mesencephalon, CER cerebellum, BST brainstem, SPC spinal cord). Expression of p150 is similar in the central nervous system (CNS), whereas that of TBCB is variable, with its highest protein levels in the PFC, CTX, and HIP (β-Actin control). d Expression profile of p150 and TBCB in subcellular compartments. Western blot was performed on pellet and soluble fractions of the postsynaptic density (PSD) preparation from 14-day-old male rats (Ho whole brain homogenate, P1 nuclei-enriched fraction, P2 membrane-associated fraction, S2 soluble fraction 2, My myelin, LM1 light membrane 1, Sy synaptosome, SJ synaptic junction). p150 was expressed throughout the subfractions with high expression in P1, My, LM1, and Sy. TBCB was expressed in all subfractions, except for the synaptic junction and PSD (β-Actin control). e, f In situ hybridization detection of DCTN1 and TBCB mRNA levels in rat brain. DCTN1 mRNA expression was high throughout developmental stages in the cortex, cerebellum, dentate gyrus, and hippocampus. Whole-mount embryonic day 19 (E19) in situ embryo shows abundant DCTN1 in the CNS (e). On the other hand, TBCB mRNA expression appears higher than DCTN1. Ubiquitous TBCB mRNA expression is seen in all organs in the whole-mount E19 embryo in situ specimen (f). In situ brain sections were obtained from E19 embryos, postnatal days 3 (PD3) and 21 (PD21), and 3-month-old (3mnth) and adult animals
p150 and TBCB are direct protein-protein interaction partners
In an initial yeast-two-hybrid screen, we used a truncated WT human p150, coding for the microtubule-binding domain, ranging from exon 1–7 as bait on a human fetal brain library. A positive clone exhibiting the full sequence of TBCB was found as a putative interaction partner (Fig. 2a). We could confirm a direct protein-protein interaction by pull-down assays and subsequent Western blotting using protein lysates of cells overexpressing either TBCB or p150 (Fig. 3e, f).
p150 and TBCB interaction, localization, and TBCB knockdown efficiency. a Yeast--two-hybrid screen. The human DCTN1 exon 1-7 gene (harboring the CAP-Gly and microtubule [MT]-binding domain, red rectangle) was used as bait on a the human male fetal cDNA library. A full-length TBCB was identified as a novel putative interacting partner. Intracellular localization of overexpressed TBCB and p150. b, d Overexpression of wild-type (WT) DCTN1 led to the localization of p150-GFP at MTs. The WT p150-GFP construct (green) was transfected into COS7 cells (b) and primary hippocampal neurons (pHNs; d) for 48 h and immunostained for alpha tubulin (red) to visualize the MT network. Upon transfection, WT DCTN1-GFP was overexpressed as a GFP fusion protein and localized to MTs. c, e TBCB overexpression was distributed throughout the cell. Likewise, TBCB was expressed either as a myc-tagged (red) or as a GFP-fusion (green) protein in pHNs (e) and COS7 (c) cells, respectively. With both plasmids, a cytoplasmic distribution of TBCB was observed in both cell types, and TBCB was also localized in all neuronal processes or neurites. Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI, blue). Bars 20 μm. f Western blot detection of endogenous TBCB and p150 in COS7 and HEK293T cell lines; 10 μg protein was loaded per lane in all experiments
a, b Assessment of knockdown of endogenous and exogenous TBCB. Downregulation of TBCB was aimed at investigating its role in neuronal development. We tested the effectiveness of two of our individual knockdown constructs (a). We overexpressed human TBCB-myc either alone or together with two TBCB RNA-interferring (RNAi1, RNAi3) constructs for 72 h aimed at a synergistic effect in HEK293T cells (b). The TBCB-overexpressed lane (a Transf.) showed a double band of overexpressed TBCB-myc above and endogenous (end.) TBCB below. The cotransfected lanes of TBCB and RNAi constructs showed a clear downregulation of both endogenous and exogenous (not shown) TBCB (b), demonstrating a positive knockdown effect (untransf. untransfected control, scrambled scrambled vector lysate control). c, d TBCB knockdown is time-dependent. Endogenous TBCB shows a time-dependent downregulation following cotransfection of both RNAi constructs (pooled together) for 48 h (c) and 72 h (d) in HEK293T cells when compared with controls (untransfected and scrambled vector lysates). e, f Immunoprecipitation experiments demonstrating p150 interaction with TBCB. Based on a yeast-two-hybrid screen, we identified TBCB as an interacting protein partner of p150Glued and confirmed this in pull-down experiments. The WT p150-GFP construct was cotransfected into HEK293T cells for 36 h with WT TBCB-myc. The cotransfected lysate was coupled to anti-myc-coated magnetic beads, and the complex was immobilized via a magnetic column in a magnetic field. The final eluate was probed with anti-GFP antibody for p150 in the precipitated complex (IP, e). The WT p150 construct and the G59S (p150 mutation involved in amyotrophic lateral sclerosis pathogenesis) constructs were precipitated in the complex when compared with the untransfected and input control lysates (f). This interaction was also seen when pulling-down the complex with mutant DCTN1 GFP constructs the other way round by detecting TBCB via the myc tag (f). g, h TBCB recruits p150Glued into its cellular compartment. To determine which of the interacting partners was recruiting the other into its compartment, we co-expressed WT p150-GFP (green) and WT TBCB-myc in both COS7 cells (g) and hippocampal neurons (h) for 24 h and subsequently immunostained for myc (red) and alpha tubulin (magenta). We observed that TBCB was able to recruit p150, giving p150 a more diffused distribution pattern (g, h). This recruiting effect was evident in COS7 cells at longer expression time points in which p150 filaments were completely absorbed (Fig. S2), whereas TBCB localization remained unaffected. This recruiting phenomenon was true for both COS7 and primary hippocampal neurons. Colocalization of both proteins was observed in both cell types, as clearly demonstrated in the inset. Cell nuclei were counterstained with DAPI (blue). Bar 20 μm
Overexpressed dynactin p150 and TBCB show different intracellular localization
The p150 protein is closely associated with the macromolecular dynactin motor complex and with the microtubule system, whereas TBCB has been reported to be localized throughout the cytoplasm (Kortazar et al. 2007). Constructs from human sequences were cloned to express either GFP or myc-tag fusion proteins. Additionally, murine p150 constructs expressing untagged p150 protein from a bicistronic mRNA, which additionally expressed a nuclear GFP were generated. Transfection of plasmids harboring DCTN1 in either COS7 cells or pHNs resulted in the overexpression of p150 proteins, displaying a clear microtubular localization, as shown by alpha tubulin counterstaining in cells (Fig. 2b, d; Electronic Supplementary Material, Fig. S2a). Overexpressed TBCB proteins showed a cytoplasmic distribution throughout the cells, whereas p150 alone was localized to the tubulin network (Fig. 2c, e). Additionally, the cell lines that we utilized throughout the studies, namely COS7 and HEK293T, were found to express endogenous TBCB and p150 (Fig. 2f). To test the overexpression and knockdown constructs of TBCB, we performed Western blot analysis and could clearly detect the overexpressed human TBCB-fusion protein and the endogenous TBCB (Fig. 3a). Transfection of TBCB-myc together with a mix of two different RNA-interferring (RNAi) constructs resulted in a decrease of both the endogenous and the overexpressed TBCB. The knockdown abilities of the RNAi constructs were time-dependent (Fig. 3b–d).
Overexpressed TBCB leads to a decreased localization of WT p150 to the microtubule network
To evaluate whether one of the proteins recruited the interaction partner into its cellular compartment, we overexpressed both proteins in COS7 cells or pHNs. An unexpected finding was that TBCB was able to recruit p150 proteins away, at least in part, from the tubular system resulting in a more diffuse distribution of p150. This was true for COS7 and pHN cells at various timepoints after transfection (Electronic Supplementary Material, Fig. S2a, b). The localization of TBCB remained unchanged (Fig. 3g, h). To quantify the mislocalization of p150 in the presence of TBCB overexpression, we aquired confocal sections of double-transfected COS7 cells and found a significant localization of p150 in the cytoplasm compared with the single transfection of p150 proteins mostly localized at filaments at various timepoints after transfection (Electronic Supplementary Material, Fig. S2c, d, e).
TBCB and putative disease relevant mutated p150 proteins
To evaluate both protein interaction and protein localization in a putative disease-specific setting, we generated constructs harboring a well-known p150 mutation involved in amyotrophic lateral sclerosis (ALS) pathogenesis (G59S) and a p150 mutation thought to be involved in neurodegeneration pathogenesis but with low penetrance or solely as a disease supporting factor (I196V). Both constructs were cloned as human GFP fusion proteins (GFP-p150) and untagged rodent proteins with a nuclear GFP (IRES-AcGFP-nuc). Pull-down experiments revealed a positive interaction of both mutated proteins with TBCB (Fig. 3f and data not shown). Whereas the single overexpression of I196V protein led to a stable localization of the mutated p150 protein to the microtubular system, the G59S protein mainly formed large cellular clusters in the cell. Additional overexpression of TBCB led to a partial loss of microtubular localization of the mutated p150 protein I196V comparable with the unmutated p150 protein. Interestingly, the G59S was able to recruit a large proportion of TBCB protein to its clusters. These observations could be documented for various time points after transfection (Electronic Supplementary Material, Fig. S1a, b).
TBCB overexpression has no apparent effect on microtubules in COS7 cells
Although TBCB overexpression has been reported to lead to microtubule disassembly (Kortazar et al. 2007; Wang et al. 2005), we could not observe any changes in the microtubular system, even after 72 h of TBCB overexpression in both neurons and COS7 cells (Fig. 4a–d), in accordance with a recent report (Cleveland et al. 2009). Neither could we observe a change in the acetylation state of alpha tubulin, acetylation of tubulin being a hint but not a prerequisite for stable microtubules (Piperno et al. 1987), after several time points (Fig. 4e). To verify the experimental setup, we applied the microtubular destabilizer colchicine, a treatment apparently leading to a complete loss of tubular structures (Fig. 4f).
TBCB overexpression has no effect on microtubule network. a–d TBCB overexpression does not disrupt microtubules. The TBCB-myc construct (red) was transfected into COS7 cells (a, b) and hippocampal neurons (c, d) at various time points. Transfected cells were identified by myc-tag immunostaining, whereas the microtubule network was examined by staining for alpha tubulin (green). At no time point was microtubule disruption observed in both cell types. Intact microtubules were prominent after 3 days of TBCB overexpression with healthy-looking DAPI-stained nuclei (blue). e Acetylation status of microtubules following TBCB overexpression is indicative of stable microtubules. HEK293T cells were transfected with TBCB at various time points, and cell lysates were probed for acetylated alpha tubulin and normal alpha tubulin. Band sizes of the acetylated and normal alpha tubulin were comparable at various time points relative to the untransfected control cell lysate, suggesting stable microtubules after TBCB overexpression (Ac-T acetylated alpha tubulin, transf. transfected, untransf. untansfected). f Colchicine disrupts microtubules, unlike TBCB overexpression. COS7 cells after 48 h in culture were treated with 10 μm colchicine for 1 h as a potent positive control for microtubule disruption. Nuclei were stained with DAPI (blue). Colchicine-treated cells completely lost their microtubule network relative to TBCB overexpressing cells that retained intact microtubules. Bar 20 μm.
TBCB overexpression has no effect on neuronal maturation or synaptogenesis
TBCB plays a role in neurogenesis in the nervous system, the highest expression being observed at the peak of cerebral and cerebellar corticogenesis (Lopez-Fanarraga et al. 2007). To evaluate the effect of TBCB on neuronal development, we overexpressed TBCB or reduced endogenous TBCB via RNAi. The overexpressed TBCB localized in the soma and throughout the neurites of hippocampal neurons. As a primary approach for investigating neuronal development, we counted the number of dendrites at various time points during in vitro cultivation. Nevertheless, overexpression of TBCB for 72 h did not lead to a significant change of primary or secondary dendrites at days 7, 10, or 13 in vitro (Fig. 5a). Subsequently, we quantified the branching points of neurites and again found no significant changes (Fig. 5b). As a last approach, we investigated the synapse number by counting Bassoon puncta along the neurites in neurons of various ages. Once more, we saw no effect on synaptogenesis in vitro under increased TBCB expression (Fig. 5c).
TBCB overexpression has no effect on neuronal maturation. a TBCB abundance has no influence on dendrite growth. The TBCB-GFP construct was transfected for 72 h into embryonic day 19 (E19)-derived rat hippocampal neurons at various time points, after which primary dendrites and secondary dendrites were counted with the aid of mitogen-activated protein 2 (MAP2) counterstaining. No significant difference was observed between the TBCB overexpressed neurons relative to the untransfected control. b Overexpressing TBCB has no effect on dendritic branching. TBCB was transfected for 72 h into E19-derived rat hippocampal neurons, and the branching points were counted for all three experimental groups with the aid of MAP2. No significant difference in branching points was observed between the TBCB overexpressing neurons compared with the untransfected control group at the stipulated time points in culture. c TBCB has no effect on synapse formation. The TBCB-GFP construct was transfected into rat E19-derived hippocampal neurons for 72 h, and points of presynaptic Bassoon in GFP-positive cells were counted per 10 μm dendrite length. No significant difference in Bassoon puncta was observed in the TBCB-overexpressing neurons relative to the untransfected control at various time points. Collectively, these data represent the cumulative means of three independent experiments with standard error of the mean (SEM); n≥20 cells per independent experiment. The mean values and SEM were normalized to the untransfected control for synapse puncta statistics (d day)
TBCB expression is not required for neuronal maturation or synaptogenesis
A second approach examined whether decreased TBCB levels had an influence on neuronal development or synaptogenesis. As also shown in Fig. 4, decreased levels of TBCB did not alter neuronal morphology or synapse number (Fig. 6a–c). This indicated that other, more important mechanisms are present in the neuron and affect microtubule assembly during early neuronal maturation, and that the main role of TBCB in this developmental stage might be different.
TBCB downregulation has no effect on neuronal maturation. As shown in Fig. 3a–d, we knocked down TBCB in hippocampal neurons for 72 h at variious time points in culture and counted Bassoon signals per 10 μm dendrite length, primary and secondary dendrites, and branching points from dendrites. As in Fig. 4, we observed no significant changes in dendrite number (a), branching point number (b), or synapse number (c) compared with the untransfected control groups at day (d) 7, 10, and 13 in vitro. Collectively, these data represent cumulative means of three independent experiments with SEM; n≥20 cells per independent experiment. The mean values and SEM were normalized to the untransfected control for synapse puncta statistics
Discussion
The cytoskeleton is assembled by a variety of functionally diverse structures. Being conferred with the ability to act as transport tracks or pathways for motor proteins, the cytoskeleton has to be built of rapidly changing and stable structures. The part of the cytoskeleton that is of tubular origin is a rapidly changing and plastic system, being adapted to regulatory processes and changes in the cell. The outside and inside diameters of these cylinders are 25 and 15 nm, respectively (Timasheff and Grisham 1980).
Microtubules consist of protofilaments of the subunit protein, tubulin, aligned longitudinally along the axis of the cylinder with 13 protofilaments arranged around the circumference. The microtubules have a helical appearance. Tubulin consists of two almost identical molecules, namely alpha tubulin and beta tubulin. Some microtubules, such as those found in cilia and flagella, are completely stable, and the functions of these microtubules are not linked to their assembly/disassembly. Most microtubules in eukaryotic cells, however, are labile. They can be found, for example, in mitotic spindles, as a disperse network in the cytoplasm, and in the axonal and dendritic compartments of a neuron. Microtubules serve here as both structural components within the cells and are further involved in many cellular processes, including mitosis, cytokinesis, and vesicular transport (Erickson 1975; Kirschner 1978; Timasheff and Grisham 1980).
The production of the native alpha/beta tubulin heterodimer in vitro depends on the action of cytosolic chaperonin and several protein cofactors. Such cofactors together with native tubulin act on beta tubulin-folding intermediates generated by the chaperonin to produce polymerizable tubulin heterodimers. TBCB is one of five chaperone proteins termed TBCA-TBCE and greatly enhances the efficiency of alpha tubulin folding in vitro (Tian et al. 1997). Nevertheless, TBCB has other mechanistic roles. It plays a part in positively regulating microtubule disassembly in vivo and is furthermore critically involved in axonal growth cone regulation (Kortazar et al. 2007; Lopez-Fanarraga et al. 2007). Other studies have shown that TBCB-mediated tubulin regulation is needed for microglia cytoskeleton reorganization during activation (Fanarraga et al. 2009). Phosphorylation of TBCB by p21-activated kinase 1 (pak1) leads to the polymerization of new microtubules, whereas a TBCB phosphorylation mutant inhibits polymerization (Vadlamudi et al. 2005), suggesting pak1 pathway regulation of microtubule dynamics via TBCB. Intriguingly, TBCB nitration antagonizes microtubule polymerization or regrowth (Rayala et al. 2007). Both phosphorylation and nitration processes are indicative of post-translational modification processes that exert an effect in the TBCB regulation of microtubule dynamics. Furthermore, TBCB degradation is regulated by gigaxonin, a protein implicated in human giant axonal neuropathy (Wang et al. 2005). Other members of the tubulin chaperone family have been shown to be involved in human diseases, such as TBCC in retinitis pigmentosa 2 (Schwahn et al. 1998) and TBCE in motor neuropathy (Bommel et al. 2002; Martin et al. 2002; Parvari et al. 2002). Alterations in microtubules, cell cycle arrest, and cell death have been reported following TBCA gene knockdown in mammalian cells (Nolasco et al. 2005). Collectively, these studies depict the potentials of tubulin cofactors in disease causation.
The dynactin motor complex, a multimeric protein complex, moves along the microtubules of the neuronal axon and are thought to be involved mainly in retrograde axonal transport (Burakov and Nadezhdina 2006). In the past few years, it has become clear that the dynactin subunit p150Glued (encoded by DCTN1) is crucially involved in neurodegenerative disorders, such as the ALS (Laird et al. 2008; LaMonte et al. 2002) or Perry syndrome (Farrer et al. 2009). Retrograde transport failure attributable to dynactin sequence aberrations is concluded to cause vesicular trafficking defects and to lead to intracellular protein aggregates (Levy et al. 2006). Other studies have reported a major effect of mutated p150 on the function of the dynein/dynactin motor complex in mouse models (Lai et al. 2007) together with proliferation of enlarged tertiary lysosomes and lipofuscin granules (Chevalier-Larsen et al. 2008). Interestingly, dynein/dynactin interaction has been suggested to be attenuated with aging (Kimura et al. 2007). At the molecular level, specific mutations affect regions of p150 leading to, for example, destabilization of the protein interaction domain (Ahmed et al. 2010).
We have performed a yeast-two-hybrid screen to identify new interaction partners of dynactin p150. Amongst others, we have found that TBCB directly interacts with p150 and have subsequently characterized the roles of dynactin and TBCB in the present study. TBCB has three functional domains: the N-terminal functional domain, the central coiled-coil region, and the C-terminal CAP-Gly or CLIP-170 domain (Radcliffe and Toda 2000). However, the TBCB motif that interacts with the microtubule-binding domain of p150 still remains to be determined. We have speculated that the CAP-Gly domain is involved in this binding, given that CAP-Gly domains have been reported to be associated with various protein-protein interactions (Steinmetz and Akhmanova 2008). Our aim has been to dissect the impact of the protein interaction partners p150Glued and TBCB. TBCB ubiquitous expression in mammalian organs has previously been shown at the RNA level (Lopez-Fanarraga et al. 2007). In this study, we confirm this data, at both the RNA and protein levels. A role for TBCB in neurogenesis and neuronal growth cone has been reported (Lopez-Fanarraga et al. 2007) but without any elaborate and specific definition of this role. In this light, we have attempted to investigate defined TBCB roles in CNS development by quantifying synapse, dendrite, and branching point numbers in developing neurons after TBCB overexpression and downregulation. We show that, at least in developing neurons, levels of TBCB have no influence on cell morphology, maturation, or the tubulin cytoskeleton. A distinct function of the dynactin motor complex in neuronal cell biology is neuronal retrograde axonal transport (Levy and Holzbaur 2006). Mutations in the p150 subunit of dynactin have been shown to be responsible for neuronal degeneration and cell death, leading, for example, to ALS (Katsuno et al. 2006). The tubulin-binding cofactors, on the other hand, are crucially involved in the assembly of tubulin subunits and later also for disassembly mechanisms of these structures (Kirschner 1978; Kortazar et al. 2007). Initially, we tried to elucidate the dual role of these proteins and overexpressed both constructs at the same time. We found that TBCB was able to modulate the subcellular localization of p150. We also observed that, under conditions of abundance, TBCB was potentially able to deplete p150 filament localization. Nevertheless, we have been unable to observe severe cellular changes or even cell death.
Interestingly, we have found that TBCB is able to recruit tubulin-bound WT p150 to its localization. This might have an influence on the long-term survival of cells under conditions in which fast and efficient changes of the tubulin cytoskeleton are required. Nevertheless, the well-known p150 mutation G59S is able to recruit TBCB to its clusters, whereas another p150 mutation, I196V does not show differences compared with the unmutated protein. Functional assembly of cytoskeletal structures and motor proteins are most important for a variety of cellular mechanisms, such as mitochondria or protein transportation. In our studies, we have not observed a major modulation of microtubule organization in our cell lines and primary neurons by the up- or downregulation of TBCB protein levels. This suggests different and still unknown roles of TBCB or the possible compensation of its deregulation via other mechanisms, including disease-specific conditions.
References
Ahmed S, Sun S, Siglin AE, Polenova T, Williams JC (2010) Disease-associated mutations in the p150(Glued) subunit destabilize the CAP-gly domain. Biochemistry 49:5083–5085
Bommel H, Xie G, Rossoll W, Wiese S, Jablonka S, Boehm T, Sendtner M (2002) Missense mutation in the tubulin-specific chaperone E (Tbce) gene in the mouse mutant progressive motor neuronopathy, a model of human motoneuron disease. J Cell Biol 159:563–569
Burakov AV, Nadezhdina ES (2006) Dynein and dynactin as organizers of the system of cell microtubules. Ontogenez 37:323–339
Chevalier-Larsen ES, Wallace KE, Pennise CR, Holzbaur EL (2008) Lysosomal proliferation and distal degeneration in motor neurons expressing the G59S mutation in the p150Glued subunit of dynactin. Hum Mol Genet 17:1946–1955
Cleveland DW, Yamanaka K, Bomont P (2009) Gigaxonin controls vimentin organization through a tubulin chaperone-independent pathway. Hum Mol Genet 18:1384–1394
Erickson HP (1975) The structure and assembly of microtubules. Ann N Y Acad Sci 253:60–77
Fanarraga ML, Villegas JC, Carranza G, Castano R, Zabala JC (2009) Tubulin cofactor B regulates microtubule densities during microglia transition to the reactive states. Exp Cell Res 315:535–541
Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, Tsuboi Y, Yamada T, Gutmann L, Elibol B, Bhatia KP, Wider C, Vilarino-Guell C, Ross OA, Brown LA, Castanedes-Casey M, Dickson DW, Wszolek ZK (2009) DCTN1 mutations in Perry syndrome. Nat Genet 41:163–165
Grabrucker A, Vaida B, Bockmann J, Boeckers TM (2009) Synaptogenesis of hippocampal neurons in primary cell culture. Cell Tissue Res 338:333–341
Katsuno M, Adachi H, Minamiyama M, Waza M, Tokui K, Banno H, Suzuki K, Onoda Y, Tanaka F, Doyu M, Sobue G (2006) Reversible disruption of dynactin 1-mediated retrograde axonal transport in polyglutamine-induced motor neuron degeneration. J Neurosci 26:12106–12117
Kimura N, Imamura O, Ono F, Terao K (2007) Aging attenuates dynactin-dynein interaction: down-regulation of dynein causes accumulation of endogenous tau and amyloid precursor protein in human neuroblastoma cells. J Neurosci Res 85:2909–2916
Kirschner MW (1978) Microtubule assembly and nucleation. Int Rev Cytol 54:1–71
Kleger A, Seufferlein T, Malan D, Tischendorf M, Storch A, Wolheim A, Latz S, Protze S, Porzner M, Proepper C, Brunner C, Katz SF, Varma Pusapati G, Bullinger L, Franz WM, Koehntop R, Giehl K, Spyrantis A, Wittekindt O, Lin Q, Zenke M, Fleischmann BK, Wartenberg M, Wobus AM, Boeckers TM, Liebau S (2010) Modulation of calcium-activated potassium channels induces cardiogenesis of pluripotent stem cells and enrichment of pacemaker-like cells. Circulation 122:1823–1836
Kortazar D, Fanarraga ML, Carranza G, Bellido J, Villegas JC, Avila J, Zabala JC (2007) Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp Cell Res 313:425–436
Lai C, Lin X, Chandran J, Shim H, Yang WJ, Cai H (2007) The G59S mutation in p150(glued) causes dysfunction of dynactin in mice. J Neurosci 27:13982–13990
Laird FM, Farah MH, Ackerley S, Hoke A, Maragakis N, Rothstein JD, Griffin J, Price DL, Martin LJ, Wong PC (2008) Motor neuron disease occurring in a mutant dynactin mouse model is characterized by defects in vesicular trafficking. J Neurosci 28:1997–2005
LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL (2002) Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron 34:715–727
Levy JR, Holzbaur EL (2006) Cytoplasmic dynein/dynactin function and dysfunction in motor neurons. Int J Dev Neurosci 24:103–111
Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, LaMonte BH, Harmison GG, Puls I, Fischbeck KH, Holzbaur EL (2006) A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J Cell Biol 172:733–745
Liebau S, Proepper C, Schmidt T, Schoen M, Bockmann J, Boeckers TM (2009) ProSAPiP2, a novel postsynaptic density protein that interacts with ProSAP2/Shank3. Biochem Biophys Res Commun 385:460–465
Liebau S, Steinestel J, Linta L, Kleger A, Storch A, Schoen M, Steinestel K, Proepper C, Bockmann J, Schmeisser MJ, Boeckers TM (2011) An SK3 channel/nWASP/Abi-1 complex is involved in early neurogenesis. PLoS One 6:e18148
Linta L, Stockmann M, Kleinhans KN, Bockers A, Storch A, Zaehres H, Lin Q, Barbi G, Bockers TM, Kleger A, Liebau S (2012) Rat embryonic fibroblasts improve reprogramming of human keratinocytes into induced pluripotent stem cells. Stem Cells Dev 21:965–976
Lopez-Fanarraga M, Carranza G, Bellido J, Kortazar D, Villegas JC, Zabala JC (2007) Tubulin cofactor B plays a role in the neuronal growth cone. J Neurochem 100:1680–1687
Martin N, Jaubert J, Gounon P, Salido E, Haase G, Szatanik M, Guenet JL (2002) A missense mutation in TBCE causes progressive motor neuronopathy in mice. Nat Genet 32:443–447
Munch C, Meyer R, Linke P, Meyer T, Ludolph AC, Haas J, Hemmer B (2007) The p150 subunit of dynactin (DCTN1) gene in multiple sclerosis. Acta Neurol Scand 116:231–234
Nolasco S, Bellido J, Goncalves J, Zabala JC, Soares H (2005) Tubulin cofactor A gene silencing in mammalian cells induces changes in microtubule cytoskeleton, cell cycle arrest and cell death. FEBS Lett 579:3515–3524
Parvari R, Hershkovitz E, Grossman N, Gorodischer R, Loeys B, Zecic A, Mortier G, Gregory S, Sharony R, Kambouris M, Sakati N, Meyer BF, Al Aqeel AI, Al Humaidan AK, Al Zanhrani F, Al Swaid A, Al Othman J, Diaz GA, Weiner R, Khan KT, Gordon R, Gelb BD (2002) Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat Genet 32:448–452
Piperno G, LeDizet M, Chang XJ (1987) Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol 104:289–302
Proepper C, Johannsen S, Liebau S, Dahl J, Vaida B, Bockmann J, Kreutz MR, Gundelfinger ED, Boeckers TM (2007) Abelson interacting protein 1 (Abi-1) is essential for dendrite morphogenesis and synapse formation. EMBO J 26:1397–1409
Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH Jr, Ludlow CL, Fischbeck KH (2003) Mutant dynactin in motor neuron disease. Nat Genet 33:455–456
Radcliffe PA, Toda T (2000) Characterisation of fission yeast alp11 mutants defines three functional domains within tubulin-folding cofactor B. Mol Gen Genet 263:752–760
Rayala SK, Martin E, Sharina IG, Molli PR, Wang X, Jacobson R, Murad F, Kumar R (2007) Dynamic interplay between nitration and phosphorylation of tubulin cofactor B in the control of microtubule dynamics. Proc Natl Acad Sci USA 104:19470–19475
Schroer TA (2004) Dynactin. Annu Rev Cell Dev Biol 20:759–779
Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, Duijnhoven G van, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, Pinckers AJ, Fundele R, Rosenthal A, Cremers FP, Ropers HH, Berger W (1998) Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19:327–332
Steinmetz MO, Akhmanova A (2008) Capturing protein tails by CAP-Gly domains. Trends Biochem Sci 33:535–545
Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ (1997) Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol 138:821–832
Timasheff SN, Grisham LM (1980) In vitro assembly of cytoplasmic microtubules. Annu Rev Biochem 49:565–591
Vadlamudi RK, Barnes CJ, Rayala S, Li F, Balasenthil S, Marcus S, Goodson HV, Sahin AA, Kumar R (2005) p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol 25:3726–3736
Vilarino-Guell C, Wider C, Soto-Ortolaza AI, Cobb SA, Kachergus JM, Keeling BH, Dachsel JC, Hulihan MM, Dickson DW, Wszolek ZK, Uitti RJ, Graff-Radford NR, Boeve BF, Josephs KA, Miller B, Boylan KB, Gwinn K, Adler CH, Aasly JO, Hentati F, Destee A, Krygowska-Wajs A, Chartier-Harlin MC, Ross OA, Rademakers R, Farrer MJ (2009) Characterization of DCTN1 genetic variability in neurodegeneration. Neurology 72:2024–2028
Wang W, Ding J, Allen E, Zhu P, Zhang L, Vogel H, Yang Y (2005) Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol 15:2050–2055
Acknowledgments
The authors thank Sabine Seltenheim and Renate Zienecker for excellent technical support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
This study was supported by the Deutsche Forschungsgemeinschaft DFG (BO1718,3-1), SFB497, B8, the Else-Kröner-Fresenius-Stiftung (2011_A200 to S.L.), EURO-MOTOR (to A.C.L. and S.L.), and Boehringer-Ingelheim BIU (N5 to S.L.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
ESM 1
(PDF 2480 kb)
Rights and permissions
About this article
Cite this article
Kuh, G.F., Stockmann, M., Meyer-Ohlendorf, M. et al. Tubulin-binding cofactor B is a direct interaction partner of the dynactin subunit p150Glued . Cell Tissue Res 350, 13–26 (2012). https://doi.org/10.1007/s00441-012-1463-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-012-1463-z