Abstract
With the rise of sequencing technologies, it is now feasible to assess the role rare variants play in the genetic contribution to complex trait variation. While some of the earlier targeted sequencing studies successfully identified rare variants of large effect, unbiased gene discovery using exome sequencing has experienced limited success for complex traits. Nevertheless, rare variant association studies have demonstrated that rare variants do contribute to phenotypic variability, but sample sizes will likely have to be even larger than those of common variant association studies to be powered for the detection of genes and loci. Large-scale sequencing efforts of tens of thousands of individuals, such as the UK10K Project and aggregation efforts such as the Exome Aggregation Consortium, have made great strides in advancing our knowledge of the landscape of rare variation, but there remain many considerations when studying rare variation in the context of complex traits. We discuss these considerations in this review, presenting a broad range of topics at a high level as an introduction to rare variant analysis in complex traits including the issues of power, study design, sample ascertainment, de novo variation, and statistical testing approaches. Ultimately, as sequencing costs continue to decline, larger sequencing studies will yield clearer insights into the biological consequence of rare mutations and may reveal which genes play a role in the etiology of complex traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
GWAS, common variants and complex traits
Complex traits such as height, type II diabetes or schizophrenia are those for which both genetics and environment contribute to the variance in the population. For most complex traits, a large number of distinct genetic loci influence the phenotypic variability. Over the past decade, genome-wide association studies (GWAS) have become the standard approach to assess the genetic contribution of complex traits. With the continued drop in genotyping costs, meta-analysis of GWAS have reached hundreds of thousands of samples enabling sufficient power to detect small effects at common single nucleotide variants [i.e., those with a minor allele frequency (MAF) ≥ 5 %]. These hypothesis-free genome-wide scans have delivered many novel discoveries, including some particularly unexpected results such as implicating the hippocampus and limbic system in BMI (Locke et al. 2015), autophagy in Crohn’s Disease (Rioux et al. 2007), and the complement system in age-related macular degeneration (Edwards et al. 2005). To date, GWAS have been used to study over 1500 traits such as post-traumatic stress disorder (Ashley-Koch et al. 2015), hoarding (Perroud et al. 2011), and type II diabetes (Replication et al. 2014) and the catalog of genome-wide significant associations contains over 23,000 variants (Welter et al. 2014).
Rare variants
Current genotyping arrays commonly used in GWAS capture most common variants through imputation, but have limited capture of variants below MAF of 5 %. With increasing sample sizes coupled with advancements in sequencing both the exome (~1 % of the genome that covers protein coding genes; [WES]) and the entire genome (WGS), the definition of rare variation has tended to shift from 5 % for the earliest GWAS to 0.5 % or even 0.1 %. Part of the initial motivation for looking at rare variants for complex traits, as opposed to Mendelian disorders, came from targeted candidate gene studies that discovered rare coding variants of large effects. For example, rare coding variants in NOD2 were linked to risk of Crohn’s Disease (Rivas et al. 2011), and rare variants in PCSK9 and ABCA1 were found to have large effects on low-density lipoprotein (LDL) cholesterol and high-density lipoprotein (HDL) levels, respectively (Cohen et al. 2004, 2005, 2006). Furthermore, successfully translating the discovery of PCSK9 to a therapeutic intervention has demonstrated the potential of taking rare variant association through to clinical application (Roth et al. 2012; Stein et al. 2012). We expect that as querying rare variants becomes increasingly feasible, their findings will continue to help identify genes and regions that contribute to the etiology of complex traits. In this review, we discuss methods for the analysis of rare variants, study design considerations, and various technologies that capture rare variation. It is intended to touch on a broad range of topics, rather than delve into specific detail; where possible we point the curious reader to additional reviews for further reading if desired.
Association testing of rare variants
Study designs
Here, we delve into the specific considerations for rare variants association studies (RVAS), covering decisions made with regards to sample ascertainment, choice of variants and statistical tests, concerns regarding population stratification, and replication. RVAS typically have one of two designs—a case–control (or cohort studies), or a family-based approach. We start by describing the analytic considerations for case–control association studies and then extend these considerations to family-based rare variant studies. For the cohort studies, we will describe the methods for case–control studies, but these methods are also largely applicable to studies of quantitative traits such as height or blood glucose levels.
Why you have to group
For rare variants, we do not observe enough copies of the minor allele to achieve sufficient levels of evidence to be convincingly associated in single marker analysis (Manolio et al. 2009). To address this issue, grouping and burden tests have long been proposed in the analysis of rare variants (Cohen et al. 2004; Li and Leal 2008; Madsen and Browning 2009; Neale et al. 2011; Neale and Sham 2004; Terwilliger and Ott 1992). These groupings aim to ensure that there are enough individuals carrying a rare variant to perform an association test. There are two main classes of group-wise tests: burden tests, where the rare variants in a region are assumed to have the same direction of effect, and variance component tests which allow for effects in opposite directions.
Burden tests
Burden tests function by comparing the number or burden of variants in cases and controls, and are the most straightforward of the gene-based tests (Li and Leal 2008; Madsen and Browning 2009; Asimit et al. 2012; Morgenthaler and Thilly 2007; Morris and Zeggini 2010). These tests collapse variants within a gene or a defined region of the genome into a single score and test for association between the score and the trait of interest. One can simply consider all variants in a gene and apply either a threshold (0 or 1) or a weight based on their functional category and/or allele frequency in the model. However, burden tests are limited by the assumption that all variants act in the same direction (i.e., all risk or all protective). Consequently, burden tests lose power if there is a mixture of both protective and risk-conferring variants in the same gene.
Variance components tests
Variance component tests (Auer and Lettre 2015; Bansal et al. 2010), most notably the sequence-based kernel association test (SKAT) (Wu et al. 2011) or C-alpha (Neale et al. 2011) (which is a special case of SKAT), were designed to address the issue in which a gene may possess a mixture of risk and protective variants. By assessing the distribution of variants, rather than their combined additive effect, these tests are robust to instances where the rare variants affect phenotype in different directions (Moutsianas et al. 2015). Thus, variance component tests are more powerful than burden tests if there is a mixture of both risk and protective variation. However, variance component tests lose power (in comparison to burden tests) when the majority of variants act in the same direction. For readers interested in a comprehensive examination of RVAS tests, see the extensive review by Lee and colleagues (Lee et al. 2014).
Which region to test
One of the central questions in RVAS, especially for WGS, is what regional definitions should be used to group rare variants in an association-testing framework. The most common choice, and arguably the most intuitive, is to aggregate variants across a gene. This is particularly appealing in exome sequencing studies where genetic variation is being measured specifically within genes. This gene-based approach can be expanded to include particular functional classes (e.g., DNase hypersensitivity sites, or all nonsense variants), all genes within a pathway, or all genes within a gene set. In the context of WGS, however, the majority of rare variants will fall outside of genes and the decision of which regions to group them over for testing becomes less clear. In this case, one could group variants by class of regulatory elements such as promoter, enhancer, or transcription factor binding site. One challenge with grouping in this manner is that regulatory elements tend to be small (100–200 bp) and thus require more samples to achieve the same power as when testing a whole gene (Zuk et al. 2014). Another way to consider aggregating rare variants, especially in the case of the noncoding region, is to use a sliding window of a specified genomic length (Psaty et al. 2009). However, determining the optimal size for a sliding window is tricky, as there is a tradeoff between using a few large windows which incurs a smaller multiple hypothesis testing burden, but comes at the cost of including variants that might be functionally unimportant or have negligible effect sizes, to using many small windows with a higher multiple testing burden. The UK10K study applied the sliding window technique with a window size of 3 kb to test 31 different traits for noncoding associations, but this analysis did not return any significant associations (The U.K.K.C. 2015).
Which variants to include
Once a specified region is chosen, one must determine which variants within that region to include in the analysis. Each individual variant will either increase the probability of having the disease (risk conferring), decrease it (protective), or have no effect on risk (neutral). Ideally, we would only include the risk-conferring variants, or alternatively only the protective variants, since including neutral variants will reduce power. However, this information is typically not known, so the challenge is to balance the chance of including the risk-conferring (or protective) variants and excluding neutral variants.
Gene level testing
When considering gene level analyses, one of the most natural approaches is to restrict to only variants predicted to truncate the protein (MacArthur et al. 2012) or ablate it through nonsense-mediated decay (Rivas et al. 2015). Four different functional categories fit in this group: frameshift, splice donor, splice acceptor, and nonsense variants. Collectively, these variants are referred to by a variety of descriptions: loss-of-function (LoF), likely gene disrupting (LGD), or protein truncating variants [PTVs (Rivas et al. 2015)]; we will use the term PTV for the remainder of this paper. One of the most attractive features of PTVs is the expectation that all the variants will act in the same direction. However, most genes in the genome are strongly conserved, meaning that natural selection keeps PTVs rare, and thus large sample sizes are necessary to observe a sufficient number of rare alleles to test for association with the trait of interest.
Another possible way to increase power without increasing sample size is to also include missense variants. However, the classification of missense variants into risk, neutral, and protective is challenging. A variety of different computational approaches for pathogenicity prediction of missense mutations have been proposed, such as SIFT (Ng and Henikoff 2001), PolyPhen2 (Adzhubei et al. 2010), MutationTaster (Schwarz et al. 2014), among others (Sunyaev 2012; Grimm et al. 2015). Each of these tools leverages different indicators of deleteriousness for missense mutations; some measure conservation [e.g., GERP++ (Davydov et al. 2010), SIFT (Ng and Henikoff 2001), phyloP (Cooper and Shendure 2011)], while others evaluate the functional effect of alternate amino acids on protein structure [PolyPhen2 (Adzhubei et al. 2010)]. Given the different sources of information of these methods, the predictions of deleteriousness often differ. Additionally, the various datasets used for training and testing these tools differ in how they define pathogenic or neutral variants, which further contributes to the inconsistency across tools (Grimm et al. 2015). We direct the reader to reviews (Grimm et al. 2015) and (Cooper and Shendure 2011) for further details regarding the variety of computational predictors of deleterious missense variants and the challenges in their utility. Regardless of the particular annotation method adopted, the resulting set of variants will likely contain a mixture of both risk and neutral variants.
Noncoding analysis
For WGS, regional definitions are considerably more challenging. Projects such as Encyclopedia of DNA Elements (ENCODE) Consortium (2012) and Epigenomics Roadmap have mapped not only genes, but also other functional elements such as promoters, enhancers, repressors, transcription factor binding sites, and methylation sites (Romanoski et al. 2015). However, many of these individual functional elements are small and unlikely to harbor sufficient numbers of rare variants for testing. Consequently, grouping together functional elements for a given gene might provide sufficient variation to perform association testing. Recently, some in silico prediction tools for assessing the deleteriousness of non-coding variants have been developed such as GWAVA (Ritchie et al. 2014), CADD (Kircher et al. 2014), and Eigen (Ionita-Laza et al. 2016). These tools provide a means to prioritize non-coding variants based on their predicted deleteriousness, in a similar fashion to what PolyPhen2 (Adzhubei et al. 2010) provides for coding variation. Such predictions can be used to define both groups of variants and the weight each variant should receive in the analysis.
For WES and WGS, a key element for selecting which variants to include and what weights to assign is to leverage frequency information. Such information can be incorporated from the sample being analyzed, as proposed in Madsen and Browning (Madsen and Browning 2009), or from a diverse population reference sample. Recently, the Exome Aggregation Consortium (ExAC) (Lek et al. 2015) has made all variants from 60,706 exomes publically available, creating an unparalleled opportunity to interrogate rare coding variants. Not only is the sample size of ExAC almost an order of magnitude larger than what was previously the biggest reference database, the NHLBI Exome Sequencing Project (N = 6515) (Fu et al. 2013), but the genetic diversity of ExAC provides a better representation of rare variants across a variety of ancestries. Leveraging external frequency information has the potential to restrict case–control analysis to extremely rare variation.
Population stratification
For case–control and cohort association studies, population stratification is a major source of type I error (Lander and Schork 1994; Pritchard and Donnelly 2001; Knowler et al. 1988); principle components analysis (PCA) and linear mixed models (LMMs) have been applied with great success in correcting for these confounders (Price et al. 2006). PCA-based correction assumes a smooth distribution of MAF over ancestry or geographical space, which is appropriate in the space of common variation. However, this approach is not appropriate for rare variation as the MAFs may be sharply localized and geographically clustered due to the fact that they have recently arisen, thus violating this assumption (Mathieson and McVean 2012). One proposed method to correct for stratification in RVAS is Fast-LLM-Select (Listgarten et al. 2013), which performs feature selection on the variants, retaining only those that are phenotypically informative to use in constructing the generalized relationship matrix (GRM). Nevertheless, Fast-LLM-Select loses power when causal variants are geographically clustered (Listgarten et al. 2013; Mathieson and McVean 2013).
Family studies
The tests described above focus mainly on case-control sequencing studies. An alternate approach is to use family-based studies including trios (i.e., father, mother, and child) and/or pedigree studies. Pedigree studies may provide a cost-effective way to capture rare variation through familial imputation as well as providing opportunities to aid in the interpretation of rare variants. For family-based studies, two main analytic approaches are available: de novo (i.e., newly arising mutations) and within-family tests, such as the transmission disequilibrium test. Here we describe the analytic considerations of these two components.
De novo tests
Studying de novo mutations is most effective under scenarios in which the selective pressure against mutations is extremely strong and the effect size for those de novo variants is large. Strong selective pressure means that when mutations arise they are rapidly removed from the population, keeping the frequency of those mutations in the population extremely low.
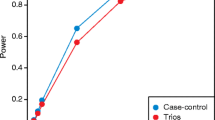
The key to analyzing de novo variation is to understand the mutability of each potential mutation site. The mutation rate across the genome varies due to a variety of factors including, but not limited to, local base context (Coulondre et al. 1978; Samocha et al. 2014), replication timing (Hardison et al. 2003; Hellmann et al. 2005; Lercher and Hurst 2002), and other large-scale phenomena (Ellegren et al. 2003). While the chance of mutation at any one gene is extremely rare (typically 2 × 10−4), across the genome we are all expected to carry ~75–100 de novo variants on average (Conrad et al. 2011; Kondrashov 2003; Vogel and Rathenberg 1975). Thus to have sufficient power to test such de novo variants for association, very large sample sizes would be required. To illustrate, ~100,000 samples are required to detect a gene in which de novo PTVs confer a 20-fold increase in risk (Zuk et al. 2014). Building a mutation rate model for de novo mutation analysis dramatically improves the power to detect genes.
Studying de novo variation for gene discovery has proved very successful for genes with large effect sizes for traits under heavy selection such as ASD (Samocha et al. 2014; De Rubeis et al. 2014; Iossifov et al. 2012, 2014; Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012), intellectual disability (de Ligt et al. 2012), developmental delay (Deciphering Developmental Disorders S 2015). An early example of this was seen in a study of Achondroplasia, in which 153 out of 154 patients had the exact same de novo variant at a CpG site in FGFR3 (Bellus et al. 1995). De novo variants have also implicated more than ten genes in ASD (De Rubeis et al. 2014; Iossifov et al. 2014) through the observation of multiple de novo PTVs in the same gene. For example, seven de novo PTVs in CHD8 have been observed in 3871 cases, a highly significant enrichment (P = 5.51 × 10−13) compared to the 0.06 that would be expected based on the mutation rate. Similar results were observed in ARID1B, SYNGAP1, DYRK1A, and other genes (De Rubeis et al. 2014).
TDT
In addition to de novo variation, rare standing variation can be analyzed for family study designs. The most commonly used association test in family designs (He et al. 2014) is the transmission disequilibrium test (TDT) (Spielman et al. 1993). The TDT can be thought of as a family-based case–control association procedure, in which the control is not a random unaffected individual but the alleles the affected child could have inherited but did not (a pseudo-control). The TDT boils down to testing whether the frequency of transmitted alleles (case) is the same as alleles not transmitted to the affected child (control) from a heterozygous parent. Because a parent who is homozygous for the variant must transmit the allele, their transmission is guaranteed and thus uninformative to the test.
Arguably the greatest advantage of the TDT is that it is free from population stratification, as the control (i.e., the untransmitted allele) is sampled from within the same family as the case. The TDT assumes Mendelian inheritance (i.e., that each allele is equally likely to be transmitted), and that a variant more often transmitted than not to the affected offspring indicates a disease-associated locus that is linked with the marker. Thus, both linkage and association are required to reject the null hypothesis; this dual hypothesis shields the TDT from population stratification. A recent study by Elansary and colleagues found that the TDT can produce false-positive associations with X-linked variants near the pseudo-autosomal region for traits with sex-limited expression and when the allele frequencies of the locus differs between the X and Y chromosomes. These false-positive associations arise because transmission is not equally likely in both sexes: fathers transmit the Y allele to their sons and the X allele to their daughters. These false positives can be fixed by considering only maternal transmissions and removing trios in which the father and mother are both heterozygous at these sites (Elansary et al. 2015).
TADA
Thus far, in focusing on only cases/control, inherited, or de novo variation, all of the association study designs discussed have utilized only partial information that can be gained from a sequencing study. This is especially true for trio-based studies, where both inherited and de novo variation can be cataloged. When multiple forms of data are available, combining them can increase power to detect association and allow for a more complete interrogation of potential disease loci. TADA (transmission and de novo association) (He et al. 2013) was developed to address this issue and integrates de novo, transmitted, and case–control variation into a unified Bayesian statistic that maximizes power to detect risk-associated genes. In terms of gene discovery, the advantage of TADA compared to using solely de novo variation scales exponentially with increasing sample size. At a sample size of 5000 trios, TADA has close to five times the power to identify associated genes compared to using only de novo variants (He et al. 2013). TADA has accelerated the pace of gene discovery in ASD, identifying 33 and 107 genes with a FDR <0.1 and <0.3, respectively (De Rubeis et al. 2014).
Additional design and analytic issues
Here we turn our attention to a range of additional issues inherent in conducting association analyses of complex traits. These issues include the relative benefits of exome versus genome sequencing, statistical considerations such as the asymptotic properties of the association tests (which relate to statistical power) as well as approaches to boosting power such as extreme phenotypic selection or the value of bottleneck populations.
Exome vs. genome
Briefly, NGS works by shearing the genome into billions of short sequence reads and aligning them to the human reference genome. Locations where the sequence differs from the reference genome are called variants. Consistent with previous reports, population-based whole genome sequencing (WGS) studies such as the 1000 Genomes (Genomes Project, C. et al. 2010) and UK10K Project (The U.K.K.C. 2015) have verified that most variants are rare. What is more, at current sample sizes the majority of variants are singletons, meaning that only one copy of the minor allele is observed in the entire sample (Fig. 1). Beyond capturing SNPs, NGS technologies also capture insertions/deletions (indels) of nucleotides, as well as more complicated structural variation such as copy-number variants (CNVs) and large-scale inversions or deletions. Current sequencing technologies capture almost all SNPs, but accurate detection of indels and structural variants still poses a challenge.
Allele frequency spectrum from exome sequencing of 2883 individuals of Swedish ancestry. The allele frequency spectrum of a typical exome sequencing study (N = 2883). The vast majority of variants are rare (MAF < 0.1 %) with 53 % being seen only once. The inset figure expands out the fraction of variants seen at allele counts 1–10
With the falling cost of WGS, rare variants are now being included in large association studies, allowing researchers to ask what role they play in complex phenotypes. While WGS is a powerful approach that enables the unbiased survey of genetic variants located genome wide, it has two main limitations. First, the costs of sequencing are still considerable, resulting in smaller samples for any one study. Second, as described above, interpreting the functional consequences of non-coding variants remains an ongoing challenge. Nevertheless, as costs continue to decline and technologies improve, WGS will likely be the standard approach for genetic investigation. However, the single most important factor in driving discovery in genetic studies is sample size, meaning that more cost-effective approaches for large samples may successfully identify significant loci more rapidly.
In contrast to WGS, whole exome sequencing (WES) targets the capture of the protein coding regions (~1.5 % of the genome). While WES is more expensive than genotyping arrays, it remains considerably less expensive than WGS. This cost-reduction enables larger sample sizes and, therefore, higher powered studies. Furthermore, our ability to interpret the functional impact of coding variants far outstrips our understanding of noncoding variation, meaning that extracting biological insight is much more straightforward (although not without its challenges). All together, these properties of the coding region increase power to identify novel associations as well as provide a better interpretation of those associations. Nevertheless, WGS projects likely have a longer shelf life than WES projects.
Extreme phenotyping
Regardless of the chosen study design, strategic choices in sample ascertainment can improve power to detect true genetic associations. This is especially important as one of the main challenges confronting RVAS is simply capturing enough rare variants to achieve sufficient observations for testing. Thus, to increase the probability that the sampled individuals will have the rare variants of interest, one popular approach is to study individuals with extreme presentations of the trait of interest (Zuk et al. 2014). The intuition behind this is that individuals at the tails of the distribution have a higher load of variants than someone in the middle. For quantitative traits, focusing on the tails of the phenotypic distribution can improve power to detect rare variant effects (Guey et al. 2011; Kryukov et al. 2009). For example, if studying the genetic drivers of height, one might gather individuals who are either very tall or very short. One can apply the same methodology to binary traits by sampling individuals with early onset of the disease. For example, an exome sequencing study of early-onset cases of chronic P. aeruginosa infection and older individuals who had not suffered infection leads to the implication of the DCTN4 in infection risk in cystic fibrosis (Emond et al. 2012).
Isolated populations and consanguineous families
Another sample ascertainment strategy is to study populations that have undergone population bottlenecks while remaining isolated for many generations (Helgason et al. 2000, 2001). These extreme bottlenecks and continued isolation (especially if followed by rapid population growth, such as in Finland) create a unique population to focus on the effects of rare variants on health. Isolated populations often have elevated allele frequencies for rare variants compared to other populations that have not experienced such events due to reduced genetic diversity from the bottlenecks and increased genetic drift from the isolation (Hatzikotoulas et al. 2014). Furthermore, population isolation results in substantial cultural and environmental homogeneity, which further increases power to find genetic factors (Auer and Lettre 2015; Hatzikotoulas et al. 2014). Restrictive and consanguineous marriage practices also produce a similar effect of elevated frequencies of variants that are rare in most other populations.
Study designs that target isolated populations have resulted in numerous successful findings. A recent study in Iceland discovered a low-frequency, non-coding variant associated with prostate cancer that was considerably more common in Iceland than in the Spanish replication cohort (3 % in cases and 1 % in controls in Iceland versus 0.4 % in cases and 0.1 % in controls in Spain) (Gudmundsson et al. 2012). Similar success, also using Icelandic individuals, has been seen for T2D (Steinthorsdottir et al. 2014). One of the most famous examples of discovery in a consanguineous group was that of BRCA1 and BRCA2 in breast and ovarian cancer in individuals of Ashkenazi Jewish descent (Levy-Lahad et al. 1997).
Asymptotics and multiple hypothesis testing
Exome sequencing has enabled RVAS to progress from candidate gene studies, where a particular gene is of interest a priori, to unbiased analyses that consider all genes in the genome. When testing all ~20,000 genes in the genome it is critical to account for multiple testing. Under the same logic of Lander and Kruglyak, given that we can test for all genes, we ought to correct for doing so (Lander and Kruglyak 1995). A Bonferroni correction for all genes brings the p value threshold required for statistical significance to 2.5 × 10−6 per gene. However, this assumes a single testing framework, which in practice is not realistic as tests of PTVs and missense mutations, either jointly or separately, are going to be conducted. Consequently, it is important to account for the diverse set of tests in such a framework, to ensure that identified associations are robust. Another possibility to correct for false discoveries in multiple tests is to use permutation. Permutations are less stringent than Bonferroni in controlling type I error rates, but can suffer from confounding when improperly done such as permuting case–control labels when the cases and controls are not ethnically matched or permuting individual genotypes (rather than phenotypes), which would fail to control for linkage disequilibrium (Kiezun et al. 2012). The use of permutation, however, can capture the total testing burden performed from the different analytic choices.
A related consideration is whether there is sufficient variation in each gene to achieve dimensionality (i.e., whether there are enough carriers of minor alleles to perform a statistical test). One way to evaluate this was proposed by Kiezun and colleagues where the data being analyzed is used to calculate what they term the i-stat, an estimate of the minimum p value achievable for a gene (Kiezun et al. 2012). Applying a threshold on i-stat-can aid in evaluating whether the gene tests are well distributed.
Factors influencing replication strategies for rare variant discovery
One of the lessons learned from GWAS was the standardization of statistical evidence required for association to avoid the failures of replication that plagued GWAS in the early years. Any GWAS now requires an initial association of P < 5 × 10−8 and independent replication for findings to be published (Barsh et al. 2012). Such standardization is necessary in the realm of RVAS as well. However, replication is far more difficult due to the fact that rare variants are by definition rare, and many times are specific to certain populations and geographic regions. Despite these difficulties, there are proposed replication strategies for whether it is a single variant, or a gene, that is being implicated.
For the former, much as in a GWAS, a cohort independent of the one in which the variant was discovered is sampled for the replication stage of the study. There are then three strategies to get at the discovered locus: directly genotype the associated variant, genotype a SNP in LD with the variant, or impute the associated variant (this being the least ideal). If the association is significant in the replication cohort, then the replication is successful.
The latter, more common design is to aggregate multiple rare variants across a gene and to test whether the gene is associated with the trait of interest. Liu and Leal describe the different ways one can go about replication in this case. Briefly, one can either resequence the gene or genotype each of the variants initially discovered in the gene in an independent population. Under the assumption that everything is equal (e.g., cost and error rates), they demonstrate that resequencing is consistently more powerful than genotyping across a number of scenarios (Liu and Leal 2010). One of the advantages of resequencing the gene is that it allows for the discovery of additional rare variants that were not present in the initial cohort. Yet in reality, genotyping and resequencing are not equal in terms of cost or accuracy. As sample size is the most important determinant of power in replication, whatever method provides the most samples would be the ideal approach (Auer and Lettre 2015).
As was the case with GWAS, we expect that continuing meta-analysis of rare variant association studies will eventually yield robust associations that continue to strengthen in significance as additional data are added. Tools such as MASS (Tang and Lin 2013, 2014), MetaSKAT (Lee et al. 2013), RAREMETAL (Liu et al. 2014; Feng et al. 2014), and seqMeta (Chen et al. 2014) have been designed to facilitate meta-analyses of rare variant association studies using summary statistics. In general, rare variant meta-analyses go through two steps: (1) calculate study-specific summary statistics, and (2) combine the summary statistics in the specified gene level association test. However, because rare variants tend to be population specific (i.e., present in only some populations), and the association analysis of these variants is conducted at the gene level, different populations and studies will contain different sets of rare variants within each gene. As a consequence, the per-study effect sizes for the gene will differ. This effect is compounded by any differences in the sequencing technology used across sites. For example, different exome capture technologies vary with respect to the efficiency of capture across different portions of the exome. Further complicating meta-analysis of rare variants is the observation that depending on the genetic architecture, a fixed-effects or random-effects model can be more powerful (Tang and Lin 2015). Of course, this is less of an issue when one is testing true loss-of-function variants in aggregate, as they should theoretically have similar or the same effect size within the same gene.
Extensions
Pathway/gene set
A natural extension of grouping rare variants together is to extend from genes to gene sets or pathways. Such tests may boost power to detect association, but necessitates accurate models of pathways and gene sets. Furthermore, the interpretation of gene set analyses can be challenging given that many gene sets tend to overlap. Nevertheless, a recent RVAS of schizophrenia (Purcell et al. 2014) reported three significant findings all with an odds ratio >5 using pathways in a cohort of 2536 cases and 2543 controls: ARC complex genes, PSD-95 complex, and voltage-gated calcium ion channel genes. Taken together, these results, along with their lack of signal at the level of single genes, suggest a polygenic architecture for schizophrenia, in which rare, disruptive variants contribute to risk (Purcell et al. 2014).
One of the complications with gene sets is that the background frequency of mutation is not the same across all genes (Samocha et al. 2014). Thus, genes with a higher rate of mutation (because of length and/or mutability) will contribute more heavily to the test statistic. Furthermore, the choice of which genes to include in the gene set is another question. One possibility is to use genes that have been implicated from GWAS as it has been established that genes harbor both common and rare variants that both affect the disease [e.g., SLC30A8 in T2D (Flannick et al. 2014)]. However, rejecting the null hypothesis does not specify which gene or genes are driving the association, thus requiring additional follow-up.
Conclusion
RVAS of complex traits are beginning to identify risk genes and causal variants, building upon the findings of GWAS that pointed to broad regions of the genome contributing to risk but that did not have the resolution that can be obtained through rare variant studies. The successes of RVAS, such as those in ASD and Inflammatory Bowel Disorder, are just the beginning of exploring the role of rare variation in complex traits. With sequencing costs dropping, new analytical methods being developed, and with the creation of large reference databases of both exomes and genomes such as the Exome Aggregation Consortium (Lek et al. 2015) and the UK10K project (The U.K.K.C. 2015), our ability to query rare variants accurately and reliably is dramatically improving. We expect to see larger and more powerful rare variant association studies continue to help hone in on underlying causal variants and inform our understanding of the genetic etiology of many common traits.
References
Adzhubei IA et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Ashley-Koch AE et al (2015) Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq-Afghanistan era veterans. J Affect Disord 184:225–234
Asimit JL, Day-Williams AG, Morris AP, Zeggini E (2012) ARIEL and AMELIA: testing for an accumulation of rare variants using next-generation sequencing data. Hum Hered 73:84–94
Auer PL, Lettre G (2015) Rare variant association studies: considerations, challenges and opportunities. Genome Med 7:16
Bansal V, Libiger O, Torkamani A, Schork NJ (2010) Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet 11:773–785
Barsh GS, Copenhaver GP, Gibson G, Williams SM (2012) Guidelines for genome-wide association studies. PLoS Genet 8:e1002812
Bellus GA et al (1995) Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet 56:368–373
Chen H et al (2014) Sequence kernel association test for survival traits. Genet Epidemiol 38:191–197
Cohen JC et al (2004) Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305:869–872
Cohen J et al (2005) Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37:161–165
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH (2006) Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 354:1264–1272
Conrad DF et al (2011) Variation in genome-wide mutation rates within and between human families. Nat Genet 43:712–714
Cooper GM, Shendure J (2011) Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet 12:628–640
Coulondre C, Miller JH, Farabaugh PJ, Gilbert W (1978) Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780
Davydov EV et al (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP ++. PLoS Comput Biol 6:e1001025
Deciphering Developmental Disorders S (2015) Large-scale discovery of novel genetic causes of developmental disorders. Nature 519:223–228
de Ligt J et al (2012) Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med 367:1921–1929
De Rubeis S et al (2014) Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515:209–215
Edwards AO et al (2005) Complement factor H polymorphism and age-related macular degeneration. Science 308:421–424
Elansary M et al (2015) On the use of the transmission disequilibrium test to detect pseudo-autosomal variants affecting traits with sex-limited expression. Anim Genet 46:395–402
Ellegren H, Smith NG, Webster MT (2003) Mutation rate variation in the mammalian genome. Curr Opin Genet Dev 13:562–568
Emond MJ et al (2012) Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet 44:886–889
ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74
Feng S, Liu D, Zhan X, Wing MK, Abecasis GR (2014) RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics 30:2828–2829
Flannick J et al (2014) Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet 46:357–363
Fu W et al (2013) Analysis of 6515 exomes reveals the recent origin of most human protein-coding variants. Nature 493:216–220
Genomes Project C et al (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073
Grimm DG et al (2015) The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum Mutat 36:513–523
Gudmundsson J et al (2012) A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet 44:1326–1329
Guey LT et al (2011) Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol 35:236–246
Hardison RC et al (2003) Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res 13:13–26
Hatzikotoulas K, Gilly A, Zeggini E (2014) Using population isolates in genetic association studies. Brief Funct Genom 13:371–377
He X et al (2013) Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet 9:e1003671
He Z et al (2014) Rare-variant extensions of the transmission disequilibrium test: application to autism exome sequence data. Am J Hum Genet 94:33–46
Helgason A et al (2000) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717
Helgason A et al (2001) mtDna and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am J Hum Genet 68:723–737
Hellmann I et al (2005) Why do human diversity levels vary at a megabase scale? Genome Res 15:1222–1231
Ionita-Laza I, McCallum K, Xu B, Buxbaum JD (2016) A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 48:214–220
Iossifov I et al (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74:285–299
Iossifov I et al (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515:216–221
Kiezun A et al (2012) Exome sequencing and the genetic basis of complex traits. Nat Genet 44:623–630
Kircher M et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315
Knowler WC, Williams RC, Pettitt DJ, Steinberg AG (1988) Gm3;5,13,14 and type 2 diabetes mellitus: an association in American Indians with genetic admixture. Am J Hum Genet 43:520–526
Kondrashov AS (2003) Direct estimates of human per nucleotide mutation rates at 20 loci causing mendelian diseases. Hum Mutat 21:12–27
Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR (2009) Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A 106:3871–3876
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048
Lee S, Teslovich TM, Boehnke M, Lin X (2013) General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet 93:42–53
Lee S, Abecasis GR, Boehnke M, Lin X (2014) Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet 95:5–23
Lek M et al (2015) Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv
Lercher MJ, Hurst LD (2002) Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet 18:337–340
Levy-Lahad E et al (1997) Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet 60:1059–1067
Li B, Leal SM (2008) Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 83:311–321
Listgarten J, Lippert C, Heckerman D (2013) FaST-LMM-Select for addressing confounding from spatial structure and rare variants. Nat Genet 45:470–471
Liu DJ, Leal SM (2010) Replication strategies for rare variant complex trait association studies via next-generation sequencing. Am J Hum Genet 87:790–801
Liu DJ et al (2014) Meta-analysis of gene-level tests for rare variant association. Nat Genet 46:200–204
Locke AE et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206
MacArthur DG et al (2012) A systematic survey of loss-of-function variants in human protein-coding genes. Science 335:823–828
Madsen BE, Browning SR (2009) A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet 5:e1000384
Manolio TA et al (2009) Finding the missing heritability of complex diseases. Nature 461:747–753
Mathieson I, McVean G (2012) Differential confounding of rare and common variants in spatially structured populations. Nat Genet 44:243–246
Mathieson I, McVean G (2013) Reply to: “FaST-LMM-Select for addressing confounding from spatial structure and rare variants”. Nat Genet 45:471
Morgenthaler S, Thilly WG (2007) A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat Res, Fundam Mol Mech Mutagen 615:28–56
Morris AP, Zeggini E (2010) An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol 34:188–193
Moutsianas L et al (2015) The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet 11:e1005165
Neale BM, Sham PC (2004) The future of association studies: gene-based analysis and replication. Am J Hum Genet 75:353–362
Neale BM et al (2011) Testing for an unusual distribution of rare variants. PLoS Genet 7:e1001322
Neale BM et al (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245
Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11:863–874
O’Roak BJ et al (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250
Perroud N et al (2011) Genome-wide association study of hoarding traits. Am J Med Genet B Neuropsychiatr Genet 156:240–242
Price AL et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Pritchard JK, Donnelly P (2001) Case-control studies of association in structured or admixed populations. Theor Popul Biol 60:227–237
Psaty BM et al (2009) Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2:73–80
Purcell SM et al (2014) A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506:185–190
Replication DIG et al (2014) Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46:234–244
Rioux JD et al (2007) Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 39:596–604
Ritchie GR, Dunham I, Zeggini E, Flicek P (2014) Functional annotation of noncoding sequence variants. Nat Methods 11:294–296
Rivas MA et al (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet 43:1066–1073
Rivas MA et al (2015) Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348:666–669
Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G (2015) Epigenomics: roadmap for regulation. Nature 518:314–316
Roth EM, McKenney JM, Hanotin C, Asset G, Stein EA (2012) Atorvastatin with or without an antibody to PCSK9 in primary hypercholesterolemia. N Engl J Med 367:1891–1900
Samocha KE et al (2014) A framework for the interpretation of de novo mutation in human disease. Nat Genet 46:944–950
Sanders SJ et al (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485:237–241
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362
Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516
Stein EA et al (2012) Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med 366:1108–1118
Steinthorsdottir V et al (2014) Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet 46:294–298
Sunyaev SR (2012) Inferring causality and functional significance of human coding DNA variants. Hum Mol Genet 21:R10–R17
Tang ZZ, Lin DY (2013) MASS: meta-analysis of score statistics for sequencing studies. Bioinformatics 29:1803–1805
Tang ZZ, Lin DY (2014) Meta-analysis of sequencing studies with heterogeneous genetic associations. Genet Epidemiol 38:389–401
Tang Z-Z, Lin D-Y (2015) Meta-analysis for discovering rare-variant associations: statistical methods and software programs. Am J Hum Genet 97:35–53
Terwilliger JD, Ott J (1992) A haplotype-based ‘haplotype relative risk’ approach to detecting allelic associations. Hum Hered 42:337–346
The UKKC (2015) The UK10K project identifies rare variants in health and disease. Nature 526:82–90
Vogel F, Rathenberg R (1975) Spontaneous mutation in man. In: Harris H, Hirschhorn K (eds) Advances in human genetics. Springer US, Boston, pp 223–318
Welter D et al (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res 42:D1001–D1006
Wu MC et al (2011) Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89:82–93
Zuk O et al (2014) Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci USA 111:E455–E464
Acknowledgments
We would like to thank all members of the ATGU and the Wall lab for their insightful discussions and assistance in writing this manuscript. We also acknowledge 1R01MH101244-02.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kosmicki, J.A., Churchhouse, C.L., Rivas, M.A. et al. Discovery of rare variants for complex phenotypes. Hum Genet 135, 625–634 (2016). https://doi.org/10.1007/s00439-016-1679-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1679-1