Abstract
Despite the family aggregation of severe teenage acne, the genetic basis of this common skin condition remains unclear. We conducted a genome-wide association study (GWAS) on severe teenage acne in 928 European Americans. The SNP rs4133274 on chromosome 8q24 (72 kb upstream of MYC) revealed the most significant association with severe teenage acne (p value = 1.7 × 10−6). The variant allele of this SNP (G allele) was associated with an increased risk of severe teenage acne with odds ratio of 4.01 (95 % confidence interval = 2.37–6.82). Upon further replication, our findings suggest new genetic basis of acne and may explain the association between acne and cancer risk observed in the epidemiological studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acne is a common condition in young people and affect all to a certain degree. It occurs earlier and is more severe in those with a family history (Ballanger et al. 2006; Ghodsi et al. 2009). Since 1984, several twin studies have assessed the heritability of acne and indicated a substantial genetic influence on familial clustering (Bataille et al. 2002; Evans et al. 2005; Friedman 1984; Niermann 1958; Walton et al. 1988). However, the individual genes responsible for this high heritability remain unclear. Although candidate gene-based studies have identified a few genetic variants associated with acne in tumor necrosis factor alpha (TNF-α) (Al-Shobaili et al. 2012; Baz et al. 2008; Szabo et al. 2011), tumor necrosis factor receptor 2 (TNFR2) (Tian et al. 2010), interleukin-1A (IL1A) (Szabo et al. 2010), cytochrome P450, family 17 (CYP17) (He et al. 2006), toll-like receptor 2 (TLR2) (Tian et al. 2010) and toll-like receptor 4 (TLR4) (Grech et al. 2012), no genome-wide association studies (GWASs) have been reported on this common skin condition. To identify novel genetic variants for acne, we conducted a genome-wide association study on severe teenage acne within a U. S cohort of young females, the Nurses’ Health Study II (NHSII).
Results
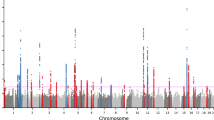
We conducted a genome-wide association study among 928 European Americans in the discovery stage (81 of them with history of severe teenage acne and 847 of them without such a history). We imputed 2,542,603 autosomal single nucleotide polymorphisms (SNPs) based on the HapMap database phase II data build 35 (CEU). After quality control filtering (see Materials and methods), 2,165,857 SNPs were selected for further association analysis with acne. The quantile–quantile (Q–Q) plots did not demonstrate a systematic deviation from the expected distribution, with the overall genomic control inflation factor (λGC) of 1.02 (Supplementary Figure 1). The Manhattan plot of each GWAS is presented in Supplementary Figure 2.
Across the whole genome, chromosome 8q24 locus showed the most significant association with severe teenage acne. In the discovery set, we identified the SNP rs4133274 on chromosome 8q24 as the SNP most significantly associated with severe teenage acne (G allele; odds ratio, OR 4.01; 95 % confidence interval, CI 2.37–6.82; P = 1.7 × 10−6, Table 1). Another SNP in the same region, rs13248513, which was in linkage disequilibrium (LD) with rs4133274 (LD r 2 = 0.85 in our study population), ranked the second (C allele, OR 3.82, 95 % CI 2.29–6.36, P = 2.2 × 10−6, Table 1). Breast cancer cases included in the discovery set might bias the results, because our previous findings suggest that severe teenage acne was associated with an increased risk of breast cancer. Hence, we further conducted a sensitivity analysis in which all breast cancer cases were excluded. The estimates of the two SNPs remained similar (OR 3.86 vs. 4.01 for rs4133274; and OR 3.86 vs. 3.82 for rs13248513). Interestingly, neither of the two SNPs was in LD with the previously identified 14 SNPs in this region that were associated with cancer risk (Grisanzio and Freedman 2010) (data not shown), and none of the cancer risk associated-SNPs was related to severe teenage acne in the present GWAS after multiple testing correction (0.05/14 = 0.004, data not shown).
We selected the top one or two loci in each chromosome region showing association with p value < 2.5 × 10−5 for a fast-track replication in an additional 1,392 European Americans from the same cohort population (134 of them with history of severe teenage acne and 1,258 of them without such a history). A total of 30 SNPs were selected for replication, but none of their associations with severe teenage acne was replicated in the replication set (Supplementary Table 1). Besides, none of the SNPs associated with acne in previous candidate gene-based studies (rs1799724 and rs1800629 in TNFα; rs1061622 in TNFR2; rs17561 in IL1A; rs743572 in CYP17; rs5743708 in TLR2; rs4986790 and rs4986791 in TLR4) (Al-Shobaili et al. 2012; Baz et al. 2008; Grech et al. 2012; He et al. 2006; Szabo et al. 2011; Tian et al. 2010) showed significant association in the GWAS of severe teenage acne (p value > 0.05; rs1799724 in TNFα and rs5743708 in TLR2 was not in our dataset).
Discussion
Our results suggested that the chromosome 8q24 locus may be associated with severe teenage acne in a GWAS among a cohort population of European Americans. Chromosome 8q24 is a large gene desert region of particular interest. This region harbors a set of risk loci associated with multiple types of cancer in previous GWASs, including prostate, breast, colon, ovarian, bladder cancers, chronic lymphocytic leukemia and glioma (Crowther-Swanepoel et al. 2010; Easton et al. 2007; Eeles et al. 2008, 2009; Ghoussaini et al. 2008; Gudmundsson et al. 2007, 2009; Shete et al. 2009; Thomas et al. 2008; Tomlinson et al. 2008; Turnbull et al. 2010; Yeager et al. 2007; Zanke et al. 2007).
Notably, we previously found that women with a history of severe teenage acne had a 17 % increased risk of breast cancer in the same cohort (data unpublished). Besides, we have previously reported a 70 % increased risk of prostate cancer among men with a history of severe acne in the Health Professionals Follow-up Study, a men’s cohort in the US (Sutcliffe et al. 2007) The closest gene around the chromosome 8q24 region is MYC, which has been known as a proto-oncogene and is a particularly compelling candidate in this region. Of interest, Myc has also been reported to regulate the androgenic effect. A Myc consensus site was identified in the androgen receptor (AR) gene to up-regulate AR (Grad et al. 1999). Previous studies also reported that Myc enhanced AR expression in androgen-independent prostate cancers and plays a key role in the control of hormone responsiveness and cell proliferation in epithelial prostatic cells (Lee et al. 2009; Silva et al. 2001). Besides, among the widespread microRNAs repressed by Myc, miR-let-7 was recently found to play an important role in the regulation of androgen signaling by down-regulating AR expression (Nadiminty et al. 2012). It is known that higher levels of circulating androgens can lead to the hyperplasia of the sebaceous glands and the seborrhea characterized by acne (Lucky 1995), and a growing body of evidence has suggested that high levels of circulating androgens are associated with increased risk of breast cancer (Eliassen and Hankinson 2008), including the NHSII (Eliassen et al. 2006). Besides, prostate cancer, a well-recognized androgen-related cancer, has been positively associated with a history of severe acne in epidemiologic study (Sutcliffe et al. 2007; Williams et al. 2012). Thus, the regulation of androgen by Myc may be a common mechanism underlying the association between acne and certain cancers. However, despite the evidence that the 8q24 region harbors regulatory elements that regulate the expression of MYC, most studies did not show a consistent association between risk allele status and MYC expression level (Grisanzio and Freedman 2010), and it cannot be ruled out that the risk variants in 8q24 may influence genes other than MYC.
One limitation of this study is the modest sample size, which may limit our statistical power to identify loci with genome-wide significance levels. Besides, we used self-reported information on acne. However, the high education level and interest in health of cohort members allow high quality and valid information to be collected on self-administered forms. In addition, a previous study demonstrated that people reporting acne of some severity were likely to have seen a physician (Cheng et al. 2010). In summary, our study suggested an association between chromosome 8q24 locus and severe teenage acne in European Americans. Upon further replication, our findings may shed new light on the genetic basis of acne and suggest a potential linkage between acne and cancer.
Materials and methods
Study population
We included 261 women with a history of severe teenage acne and 2,578 women without such a history in the NHSII cohort in the present GWAS. The NHSII is a prospective cohort study established in 1989, when 116,430 female registered nurses aged 25–42 and residing in the United States at the time of enrollment responded to an initial questionnaire on their medical histories and baseline health-related exposures. Details of this cohort have been described previously (Bertone-Johnson et al. 2009). The protocol for this study was approved by the Institutional Review Board at Brigham and Women’s Hospital and the Harvard School of Public Health.
Participants reported their history of severe teenage acne on the baseline questionnaire in 1989. In the discovery stage, we used pooled samples from two existing GWASs nested within the NHSII: one was a part of the Cancer Genetic Markers of Susceptibility project (CGEMS, n = 290; 34 with a history of severe teenage acne and 256 without such a history) and the other was a kidney stone case–control study (n = 638; 47 with a history of severe teenage acne and 591 without). All the NHSII individuals in the CGEMS set were breast cancer cases. In the replication stage, we recruited an additional 1,392 women in a renal function study nested in the same cohort for a fast-track replication (134 with a history of severe teenage acne and 1,258 individuals without such a history). We provided the detailed descriptions of each component data set in the Supplementary Material. There was no overlapping of samples among these datasets. All subjects included in this analysis were US non-Hispanic Europeans.
Genotyping, imputation and quality control
We used the Illumina 610K SNP chip for genotyping individuals in the discovery set. We imputed 2,542,603 autosomal SNPs based on haplotypes from the HapMap (http://www.hapmap.org) phase II data build 35 (CEU) (Marchini et al. 2005) using the MACH program (Li et al. 2009). Samples from the two component study sets were imputed separately. SNPs with imputation r 2 > 0.4 and minor allele frequency (MAF) >0.05 in both studies were included. Finally, 2,165,857 SNPs were included in the association analysis. The top SNPs selected from the discovery stage were genotyped in the replication set using TaqMan/BioTrove assays at the Dana Farber/Harvard Cancer Center Polymorphism Detection Core. Laboratory personnel were blinded to the case–control status, and blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100 %. Primers, probes and conditions for genotyping assays are available upon request. All these SNPs had P value for the Hardy–Weinberg test of equilibrium >0.002 (0.05/30). We excluded two SNPs rs7270170 and rs6996538 with low call rates.
Statistical analysis
Logistic regression was applied to evaluate the association between the minor allele counts and severe teenage acne. In the discovery stage, we used the imputed dosage data and adjusted for the first four principal components as well as the data set in the model. These principal components were calculated for all individuals on the basis of ca. 10,000 unlinked markers using the EIGENSTRAT software (Price et al. 2006). We used ProbABEL for the GWAS analysis and used Statistical Analysis System software (version 9.1.3; SAS Institute, Cary, NC, USA) for the replication study. Associations in the discovery set and the replication set were combined in an inverse variance-weighted meta-analysis using the METAL software (http://www.sph.umich.edu/csg/abecasis/Metal/index.html).
References
Al-Shobaili HA, Salem TA, Alzolibani AA, Robaee AA, Settin AA (2012) Tumor necrosis factor-alpha -308 G/A and interleukin 10–1082 A/G gene polymorphisms in patients with acne vulgaris. J Dermatol Sci 68:52–55
Ballanger F, Baudry P, N’Guyen JM, Khammari A, Dreno B (2006) Heredity: a prognostic factor for acne. Dermatology 212:145–149
Bataille V, Snieder H, MacGregor AJ, Sasieni P, Spector TD (2002) The influence of genetics and environmental factors in the pathogenesis of acne: a twin study of acne in women. J Invest Dermatol 119:1317–1322
Baz K, Emin Erdal M, Yazici AC, Soylemez F, Guvenc U, Tasdelen B, Ikizoglu G (2008) Association between tumor necrosis factor-alpha gene promoter polymorphism at position -308 and acne in Turkish patients. Arch Dermatol Res 300:371–376
Bertone-Johnson ER, Hankinson SE, Johnson SR, Manson JE (2009) Timing of alcohol use and the incidence of premenstrual syndrome and probable premenstrual dysphoric disorder. J Womens Health (Larchmt) 18:1945–1953
Cheng CE, Irwin B, Mauriello D, Liang L, Pappert A, Kimball AB (2010) Self-reported acne severity, treatment, and belief patterns across multiple racial and ethnic groups in adolescent students. Pediatr Dermatol 27:446–452
Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, Ruiz-Ponte C, Enjuanes A, Rosenquist R, Carracedo A, Jurlander J, Campo E, Juliusson G, Montserrat E, Smedby KE, Dyer MJ, Matutes E, Dearden C, Sunter NJ, Hall AG, Mainou-Fowler T, Jackson GH, Summerfield G, Harris RJ, Pettitt AR, Allsup DJ, Bailey JR, Pratt G, Pepper C, Fegan C, Parker A, Oscier D, Allan JM, Catovsky D, Houlston RS (2010) Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet 42:132–136
Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE et al (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447:1087–1093
Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40:316–321
Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G et al (2009) Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 41:1116–1121
Eliassen AH, Hankinson SE (2008) Endogenous hormone levels and risk of breast, endometrial and ovarian cancers: prospective studies. Adv Exp Med Biol 630:148–165
Eliassen AH, Missmer SA, Tworoger SS, Spiegelman D, Barbieri RL, Dowsett M, Hankinson SE (2006) Endogenous steroid hormone concentrations and risk of breast cancer among premenopausal women. J Natl Cancer Inst 98:1406–1415
Evans DM, Kirk KM, Nyholt DR, Novac C, Martin NG (2005) Teenage acne is influenced by genetic factors. Br J Dermatol 152:579–581
Friedman GD (1984) Twin studies of disease heritability based on medical records: application to acne vulgaris. Acta Genet Med Gemellol (Roma) 33:487–495
Ghodsi SZ, Orawa H, Zouboulis CC (2009) Prevalence, severity, and severity risk factors of acne in high school pupils: a community-based study. J Invest Dermatol 129:2136–2141
Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, Whittemore AS, Gayther SA, Giles GG, Guy M, Edwards SM, Morrison J, Donovan JL, Hamdy FC, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Brown PM, Hopper JL, Neal DE, Pharoah PD, Ponder BA, Eeles RA, Easton DF, Dunning AM (2008) Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100:962–966
Grad JM, Dai JL, Wu S, Burnstein KL (1999) Multiple androgen response elements and a Myc consensus site in the androgen receptor (AR) coding region are involved in androgen-mediated up-regulation of AR messenger RNA. Mol Endocrinol 13:1896–1911
Grech I, Giatrakou S, Damoraki G, Pistiki A, Kaldrimidis P, Giamarellos-Bourboulis EJ, Stavrianeas N (2012) Single nucleotide polymorphisms of toll-like receptor-4 protect against acne conglobate. J Eur Acad Dermatol Venereol 26:1538–1543
Grisanzio C, Freedman ML (2010) Chromosome 8q24-associated cancers and MYC. Genes Cancer 1:555–559
Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K (2007) Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet 39:631–637
Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K (2009) Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 41:1122–1126
He L, Yang Z, Yu H, Cheng B, Tang W, Dong Y, Xiao C (2006) The relationship between CYP17 -34T/C polymorphism and acne in Chinese subjects revealed by sequencing. Dermatology 212:338–342
Lee JG, Zheng R, McCafferty-Cepero JM, Burnstein KL, Nanus DM, Shen R (2009) Endothelin-1 enhances the expression of the androgen receptor via activation of the c-myc pathway in prostate cancer cells. Mol Carcinog 48:141–149
Li Y, Willer C, Sanna S, Abecasis G (2009) Genotype imputation. Annu Rev Genomics Hum Genet 10:387–406
Lucky AW (1995) Hormonal correlates of acne and hirsutism. Am J Med 98:89S–94S
Marchini J, Donnelly P, Cardon LR (2005) Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet 37:413–417
Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X, eVere White RW, Kung HJ, Evans CP, Gao AC (2012) MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem 287:1527–1537
Niermann H (1958) Report on 230 twins with skin diseases. Z Mensch Vererb Konstitutionsl 34:483–487
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El Hallani S, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS (2009) Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41:899–904
Silva IS, Morsch DM, Urnauer L, Spritzer PM (2001) Androgen-induced cell growth and c-myc expression in human non-transformed epithelial prostatic cells in primary culture. Endocr Res 27:153–169
Sutcliffe S, Giovannucci E, Isaacs WB, Willett WC, Platz EA (2007) Acne and risk of prostate cancer. Int J Cancer 121:2688–2692
Szabo K, Tax G, Kis K, Szegedi K, Teodorescu-Brinzeu DG, Dioszegi C, Koreck A, Szell M, Kemeny L (2010) Interleukin-1A +4845(G > T) polymorphism is a factor predisposing to acne vulgaris. Tissue Antigens 76:411–415
Szabo K, Tax G, Teodorescu-Brinzeu D, Koreck A, Kemeny L (2011) TNFalpha gene polymorphisms in the pathogenesis of acne vulgaris. Arch Dermatol Res 303:19–27
Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40:310–315
Tian LM, Xie HF, Yang T, Hu YH, Li J, Wang WZ (2010) Association study of tumor necrosis factor receptor type 2 M196R and toll-like receptor 2 Arg753Gln polymorphisms with acne vulgaris in a Chinese Han ethnic group. Dermatology 221:276–284
Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS (2008) A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 40:623–630
Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den Ouweland A, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, Easton DF (2010) Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet 42:504–507
Walton S, Wyatt EH, Cunliffe WJ (1988) Genetic control of sebum excretion and acne–a twin study. Br J Dermatol 118:393–396
Williams HC, Dellavalle RP, Garner S (2012) Acne vulgaris. Lancet 379:361–372
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 39:645–649
Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG (2007) Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 39:989–994
Acknowledgments
We thank Constance Chen for programming support, and thank Pati Soule and Dr. Hardeep Ranu of the Dana Farber/Harvard Cancer Center High-Throughput Polymorphism Detection Core for sample handling and genotyping of the NHSII samples. We thank the participants in the Nurses’ Health Study II for their dedication and commitment. In addition, we would like to thank the participants and staff of the Nurses’ Health Study II for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, and WY. The NHSII cohort is supported by NIH grant CA50385 and CA67262.
Conflict of interest
The authors declare that there are no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1. Quantile–Quantile (QQ) plot. QQ plot of observed vs. expected -log10(P) values for the GWAS of severe teenage acne.
Supplementary Figure 2. Manhattan plot. Manhattan plot for the GWAS of severe teenage acne.
Rights and permissions
About this article
Cite this article
Zhang, M., Qureshi, A.A., Hunter, D.J. et al. A genome-wide association study of severe teenage acne in European Americans. Hum Genet 133, 259–264 (2014). https://doi.org/10.1007/s00439-013-1374-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-013-1374-4