Abstract
One of the most severe forms of infertility in humans, caused by gametogenic failure, is non-obstructive azoospermia (NOA). Approximately, 20–30% of men with NOA may have single-gene mutations or other genetic variables that cause this disease. While a range of single-gene mutations associated with infertility has been identified in prior whole-exome sequencing (WES) studies, current insight into the precise genetic etiology of impaired human gametogenesis remains limited. In this paper, we described a proband with NOA who experienced hereditary infertility. WES analyses identified a homozygous variant in the SUN1 (Sad1 and UNC84 domain containing 1) gene [c. 663C > A: p.Tyr221X] that segregated with infertility. SUN1 encodes a LINC complex component essential for telomeric attachment and chromosomal movement. Spermatocytes with the observed mutations were incapable of repairing double-strand DNA breaks or undergoing meiosis. This loss of SUN1 functionality contributes to significant reductions in KASH5 levels within impaired chromosomal telomere attachment to the inner nuclear membrane. Overall, our results identify a potential genetic driver of NOA pathogenesis and provide fresh insight into the role of the SUN1 protein as a regulator of prophase I progression in the context of human meiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
According to estimates, male factor infertility affects 30–55% of infertile couples, with about 1 in 13 men of childbearing age needing professional care to conceive (Anderson et al. 2009). A disorder known as azoospermia, which affects an estimated 1% of males and 10–15% of male infertility cases, causes the ejaculate to be empty of sperm (Jarvi et al. 2010). The two types of azoospermia are obstructive (OA) and non-obstructive (NOA), with the latter accounting for approximately 60% of cases and resulting from primary testicular failure or secondary impairment of normal testicular function due to endocrine disorders or other conditions that impair spermatogenesis (Arshad et al. 2020).
Meiotic arrest and significantly decreased spermatogenesis are symptoms of NOA in males (Xie et al. 2022). A range of adverse factors can contribute to NOA incidence, including trauma, infection, hormonal imbalances, exposure to chemotherapy or radiotherapy, and various chromosomal abnormalities or genetic mutations (Arshad et al. 2020). Genetic defects are estimated to contribute to approximately 15% of male infertility cases and 21–29% of azoospermia incidence (Krausz and Riera-Escamilla 2018; Majzoub et al. 2022). Numerous genes have been recognized as essential for meiotic recombination, and mutations in these genes are usually found in people with NOA (Xie et al. 2022). Even so, the precise genetic mechanisms that govern gametogenic failure in this context remain incompletely understood. This study used next-generation sequencing analyses of samples from an NOA patient to identify a novel pathogenic variant in the meiosis-related SUN1 (Sad1 and UNC84 domain containing 1) gene [c. 663C > A:p.Tyr221X].
The highly controlled process of meiotic cell division produces haploid gamete cells from diploid progenitor cells. The longest meiotic stage, prophase I, includes the substages known as leptotene, zygotene, pachytene, diplotene, and diakinesis, during which the presence of chromosomes can distinguish. As homologous chromosome segregation during prophase I ultimately result in the halving of chromosome numbers in daughter cells, many different mechanisms act to regulate homologous chromosome pairing, synapsis, double-stranded DNA break (DSB) formation and repair, and bivalent remodeling (Bolcun-Filas and Handel 2018).
During prophase I, the pairing of homologous chromosomes provides an opportunity for recombination. The telomeric structures at the ends of these chromosomes attach to the nuclear envelope (NE). It transiently clusters them into a configuration known as a "bouquet" in a small area of the NE before such pairing can occur (Harper et al. 2004; Scherthan 2001). This activity is crucial for subsequent chromosomal pairing (Scherthan et al. 1996). The linker of the nucleoskeleton and cytoskeleton (LINC) complex has been identified as a critical mediator that connects the cytoskeleton and these telomeric structures (Burke 2018; Kmonickova et al. 2020) with the LINC complex components SUN1 and KASH5 (Klarsicht/ANC-1/Syne/homology 5) having been shown to, respectively, cross the inner and outer nuclear membranes (INM and ONM). While KASH5 can interact with the microtubule-associated dynein–dynactin complex (Horn et al. 2013; Morimoto et al. 2012), SUN1 facilitates telomeric tethering to the INM (Ding et al. 2007).
In addition to the LINC complex, another INM-associated complex (TTM) consisting of meiosis-specific structural molecules TERB1, TERB2 (telomere repeat-binding bouquet formation proteins 1 and 2), and MAJIN (membrane-anchored junction protein) was identified to be implicated in tethering telomeres to the INM (Shibuya et al. 2015). As a “linker” bridging LINC and the telomeres, TTM binds to telomeres through the transmembrane DNA-binding activity of MAJIN, and an interaction between TERB1 with a telomeric-shelterin protein TRF1 (Chen et al. 2021; Salas-Huetos et al. 2021). All of these proteins are critical for telomere anchoring in INM.
The knockout of either SUN1 or KASH5 in mice can interfere with the proper formation of the LINC complex, thus disrupting telomeric attachment and the movement of homologous chromosomes in a manner that contributes to gametogenic arrest and infertility in males and females that are irreversible (Ding et al. 2007; Horn et al. 2013; Morimoto et al. 2012). Multiple studies have reported variants in the KASH5 gene associated with human NOA cases (Fakhro et al. 2018; Wu et al. 2022; Zhang et al. 2022), whereas there have not been any reports of NOA resulting from SUN1 variants. Here, we sought to determine the impact of the identified human SUN1 variant on meiotic activity. Our results suggest that SUN1 functions as a regulator of human testicular function, underscoring a potential genetic driver that can contribute to the pathogenesis of NOA.
Materials and methods
Clinical samples
Study participants were recruited through the Department of Center for Reproduction and Genetics, Suzhou Municipal Hospital. NOA was diagnosed as per World Health Organization guidelines based on analyses of semen samples. Participants were not eligible for study participation if they had any history of factors known to contribute to NOA development, such as mumps, abnormal karyotypes, or genomic azoospermia factor deletions. All individuals gave written informed consent to participate in research. The proband and a single fertile individual with OA who had testicular biopsy provided testicular tissue samples. The Institutional Review Board of Suzhou Municipal Hospital, China, approved this study (No. 2019190).
Whole-exome sequencing
The sequencing of the whole exome permits the sequencing of protein-coding genes. For this investigation, WES was done on DNA samples from the proband (P06) using a TIANamp Blood DNA Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer's instructions. According to the manufacturer's instructions, exome capture and sequencing were done using the Hiseq2000 platform (Illumina, CA, USA) and AlExome Enrichment kit V1 (iGeneTech, Beijing, China). Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net/) was used to align reads to the hg19 (GRCh37) reference genome using the default parameters. The Genome Analysis Toolkit HaplotypeCaller (http://www.broadinstitute.org/gatk/) was used to call genomic variants, filtered and annotated with the ANNOVAR software (https://annovar.openbioinformatics.org/en/latest/).
Exclusion criteria included any variants with an allele frequency > 1% as determined by the ExAC Browser and 1000 Genomes Project and any variants in upstream, downstream, or intronic regions. The variants identified by SIFT, PolyPhen-2, and Mutation Taster as nonsense, frameshift, crucial splice site, or possible negative missense variants were kept for examination. Genes with two potentially detrimental missense or loss-of-function mutations were retained. Moreover, candidate genes were compared to known testis-enriched genes and genes associated with the pathogenesis of azoospermia in mice, with Sanger sequencing being used to investigate positive candidate variants.
Sanger sequencing
Using the primers specified in Supplementary Table S1, the genomic area of interest was amplified by PCR using Phanta™ Super-Fidelity DNA Polymerase (Vazyme, P501) and the following thermocycler settings: 95 ℃ for 2 min; 35 cycles of 95 ℃ for 10 s, 58 ℃ for 15 s, 72 ℃ for 8 min, and 72 ℃ for 10 min. Following direct sequencing using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit and the ABI 3100 Genetic Analyzer (Applied Biosystems, CA, USA), PCR results (359 bp) were examined through 1% agarose gel electrophoresis. SnapGene (v 3.2.1) was used to align DNA sequences.
qPCR
To extract DNA from tissue samples, an RNeasy Plus Micro Kit with on-column DNase digestion was used (Qiagen Ltd., 74034). In a sterile Teflon micropestle on ice, samples were first homogenized in 350 μL of RLT buffer with 4 μL of β-mercaptoethanol. Following sample processing according to the kit's instructions, RNA was extracted in 14 μL of RNase-free water. Then, using the PrimeScript RT reagent Kit (TaKaRa Bio Inc., RR037A) with random and oligo (dT) primers and the following thermocycler settings: 37 °C for 15 min, 85 °C for 5 s, cDNA was generated from 1 ug of RNA per sample. Samples were maintained on ice. Then qPCR analyses were performed in triplicate using SYBR Green and a StepOne-Plus instrument (Applied Biosciences) using appropriate primers (Supplementary Table S1), with 18S rRNA serving as a normalization control.
Western immunoblotting
RIPA buffer (CWBio, CW2333) supplemented with protease, and phosphatase inhibitor cocktails (CWBio, CW2200, and CW2383) were used to lyse tissue samples for 30 min on ice, followed by centrifugation for 30 min at 16,000×g. Lysates were then boiled for 5 min in a loading buffer (FDBio, FD002) after Western immunoblotting was performed, as reported previously (Zheng et al. 2015). Briefly, following 4–20% SDS-PAGE separation, proteins were transferred to PVDF membranes (Millipore, IPVH00010) that were subsequently blocked for 2 h with 5% non-fat milk at room temperature probed at 4 °C overnight with primary antibodies. Bands were identified using an enhanced chemiluminescence kit (FDBio, FD8020) after blots were rinsed in triplicate with TBST, HRP-conjugated secondary antibodies, and room temperature incubation for 2 h. Supplementary Table S2 lists the antibodies utilized in this study.
Histochemical staining
Bouin's solution was used to fix sections of human testicular tissue overnight, after which these sections were paraffin-embedded and cut into 5 μm-thick sections. The sections were then stained with hematoxylin and eosin (H&E) after being deparaffinized with xylene for 20 min, rehydrated using an ethanol gradient (100%, 90%, 80%, and 70%; 2 min each), then stained with hematoxylin for 2 min and eosin for 5 min. Sections were then dehydrated with an ethanol gradient (70%, 80%, 90%, and 100%; 2 min each) and treated for 10 min with xylene before sealing with neutral resin for analysis.
Structurally preserved nuclei preparation
Structurally preserved nuclei were prepared using a slightly modified version of a previously published protocol (Shibuya et al. 2015). Briefly, testes samples were treated for 15 min at 37 °C with trypsin–EDTA, followed by pipetting and repeated centrifugation. Pellets were then repeatedly washed with PBS before the suspension for 5 min at room temperature in a hypotonic buffer (30 mM Tris [pH7.5], 17 mM trisodium citrate, 5 mM EDTA, 50 mM Sucrose). The cell suspensions were then transferred to microscope slides in an equivalent amount of fixation buffer (1.0% PFA, 0.1% Triton X-100). After 6 h of fixing at room temperature, they were air dried under mild extraction conditions.
Chromosome spreads
An earlier reported methodology for chromosome spreads and immunofluorescent labeling was slightly modified for these procedures (Peters et al. 1997). Immediately after being minced with scalpels in DMEM containing 10% fetal calf serum, samples of control or mutant testis tissue were run through 40-μm mesh filters. After rinsing with PBS, the cells were centrifuged for 5 min at room temperature before being resuspended in 1.5 mL of hypotonic extraction buffer (30 mM Tris, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF, pH 8.2). Cells were resuspended in 200 μl of a 100 mM sucrose solution and distributed onto a thin layer of paraformaldehyde containing Triton X-100 after the supernatants were transferred to fresh tubes and centrifuged once more. This procedure was followed after a 30-min incubation at room temperature. These slides were fixated for 3 h and then air dried at ambient temperature.
Immunofluorescent staining
Samples were blocked for 1 h at room temperature with 5% BSA and probed overnight with the appropriate primary antibodies at 4 °C following three washes with PBS containing 0.1% Triton X-100 (PBST) (Supplementary Table S2). After three washes with PBST, samples were probed for one hour at room temperature with secondary antibodies, and stained for five minutes with Hoechst 33342 (Invitrogen, H21492). A confocal laser scanning microscope (LSM 800, Carl Zeiss) was then used for sample imaging, with ZEN 2012 Bule Edition (Carl Zeiss) being used for signal quantification.
Statistical analysis
GraphPad Prism 9.0 was used to analyze the data, and quantitative data were expressed as means ± SD from at least three technical replicates. Unpaired Student’s t tests were used to examine differences between P06 and a control patient with OA, with P < 0.05 regarded as the significant level.
Results
Whole-exome sequencing identifies homozygous SUN1 mutations in a patient with NOA
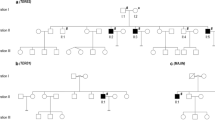
Whole-exome sequencing (WES) is a strategy that can readily aid in exploring the genetic basis for male factor infertility. It has identified many different NOA-related genetic mutations in prior studies (Gershoni et al. 2019; Yatsenko et al. 2015; Yu et al. 2021). Among a cohort of NOA patients, we identified a single patient (P06) carrying a stop codon gain variant (c.[663C > A]) in Exon 5 of SUN1 (Fig. 1A and Table 1). This variation has not been reported before in the Genome Aggregation Database. The analysis of P06's sperm samples according to the WHO guidelines revealed that despite the normal semen volume, sperm were lacking (Table 1). P06 was of average height, intelligence, and secondary sexual features. His FSH, LH, estrogen, prolactin, and testosterone levels were within normal range. He was karyotypically normal, with no signs of Y chromosome microdeletions. Ultrasonography revealed that both testicles were healthy (right: 11 mL, left: 13 mL; normal range: 10–15 mL).
Pedigree analyses and Sanger sequencing-based validation of the identified SUN1 variant. A Pedigree analysis for the sterile proband patient revealed a consanguineous family in which one individual had been diagnosed with NOA. The individual indicated with an arrow underwent WES analyses. Double horizontal lines denote consanguineous relationships. Circles and squares indicate females and males, with solid and open symbols indicating infertility and no impact on fertility. B Sanger sequencing was used to confirm the presence of the c. 663C > A SUN1 variant in family members, with the proband being homozygous for this variant while the proband's parents and uncle were heterozygous carriers. C A schematic overview of the position of the identified SUN1 variant at the genomic and protein level
A homologous frameshift variant in SUN1 results in the complete absence of SUN1 expression in spermatocytes
Sanger sequencing of SUN1 was carried out to verify the WES results using samples from P06 and available family members. This approach confirmed the co-segregation of the identified variant, as both proband's patients were heterozygous carriers for this variant (Fig. 1). Those mRNAs harbor premature stop codons typically undergo selective degradation through selective degradation, a process known as nonsense-mediated decay (Chang et al. 2007; Mühlemann 2016). When translated, these mRNAs also tend to give rise to truncated proteins that exhibit dominant negative or deleterious gain-of-function phenotypes (Chang et al. 2007). To more fully explore the potential pathogenicity of this SUN1 variant, testicular biopsy samples were obtained from P06 and a control male OA patient exhibiting normal spermatogenesis. As expected, testis samples from P06 revealed a > 80% reduction in SUN1 expression at the mRNA level as measured via qPCR (Fig. 2A). Moreover, Western immunoblotting indicated that SUN1 protein expression was undetectable in P06 samples (Fig. 2B). These data thus confirmed the absence of SUN1 expression in testes from P06.
The identified SUN1 variant contributes to a loss of SUN1 expression. A qPCR analyses of patient testis samples confirmed the reduction of SUN1 expression at the mRNA level. Data are means ± SD from three independent technical replicates. **P < 0.01. B Western immunoblotting revealed the complete absence of SUN1 protein expression in patient testis tissues. C Spermatocytes from P06 and a control patient with OA were stained with antibodies specific for lamin B and SUN1. The arrow indicates SUN1 localized at the nuclear envelope. D Spread spermatocytes from P06 and a control OA patient were stained with antibodies specific for SYCP3 and SUN1. The arrows indicate SUN1, which colocalizes with SYCP3. OA, obstructive azoospermia. Asterisk, apoptotic cells
SUN1 exhibits localization with telomeres during the leptotene and pachytene stages of meiosis
SUN1 has been shown to associate with the INM, providing a mechanism through which chromosomes can attach to the NE during meiosis via binding between SUN1 and telomeres (Ding et al. 2007). To understand the dynamic role that SUN1 plays in meiotic progression in humans, we next assessed SUN1 in spermatocytes from P06. In line with the above Western immunoblotting data, SUN1 was not detectable via immunofluorescence at the nuclear membrane in spermatocytes from this patient. In contrast, it was found at the nuclear membrane in OA control patient spermatocytes (Fig. 2C). The staining for the synaptonemal complex axial element protein SYCP3 was used to distinguish between spermatocyte stages while analyzing SUN1 in disseminated spermatocytes. SUN1 signal was visible throughout meiotic development in spermatocytes from a control OA patient, colocalizing with the synaptonemal complex axial element (Fig. 2D). This shows that SUN1 may regulate telomere attachment to the nuclear envelope in human spermatocytes, consistent with its function in a mouse model.
SUN1 is required for meiotic recombination and progression
To better understand the relationship between SUN1 expression and fertility, testicular sections were subjected to H&E staining. Relative to samples from an OA control patient, the seminiferous tubules of P06 contained spermatogonia and spermatocytes but no post-meiotic germ cells. This was indicative of spermatogenesis arrest at the spermatocyte stage in this patient with the discovered SUN1 mutation (Fig. 3A). During the progression of meiosis, SPO11 catalyzes DSB formation following the PRDM9-mediated chromatin changes at recombination hotspots. These DSBs provoke a DNA damage response, resulting in ATM kinase activation and H2AX phosphorylation (γH2AX) (Bellani et al. 2005; Parvanov et al. 2010), and staining for γH2AX can enable the assessment of spermatocyte staging. Accordingly, spermatocytes from the control OA patient and P06 were stained for SYCP3 and γH2AX, with the latter being present throughout the nucleus on autosomal chromatin at the leptotene and zygotene stage (Fig. 3B). After meiotic double-strand break (DSB) repair in normal spermatocytes, this γH2AX signal vanishes and is only detected on XY chromatin during the diplotene and pachytene stages, coinciding with the inactivation of sex chromosomes. In contrast to this normal behavior, pachytene-like SUN1-deficient spermatocytes exhibited persistent nuclear γH2AX-positivity and the absence of sex body formation (Fig. 3B), indicating that meiotic DSB repair does not occur properly in these cells (Fig. 3C).
Analysis of spermatocyte arrest at the leptotene- and zygotene-like stages. A H&E staining was used to analyze testicular samples from P06 and a male control OA patient. P, pachytene spermatocyte; RS, round spermatid; ES, elongated spermatid. B Immunofluorescent staining was used to evaluate surface-spread spermatocytes from P06 and a control OA patient with staining using antibodies specific for SYCP3 (green) and γH2AX (red). The arrowhead indicates γH2AX on the sex chromosome; The hollow arrowhead indicates γH2AX trapped on the autosomes. C Spermatocyte population composition in samples from P06 and the control OA patient, with numbers of analyzed spermatocytes (n) provided in parentheses. L leptotene, Z zygotene, P-like pachytene-like, P pachytene, D diplotene. Data are means ± SD from three independent technical replicates. **P < 0.01; ****P < 0.0001
Human spermatocytes deficient for SUN1 expression exhibit impaired telomere attachment to the NE
Telomeres are key mediators of meiotic prophase I, although the mechanisms underlying this relationship require further clarification (Mikolcevic et al. 2016; Scherthan 2007). SUN1 can interact with telomeric regions of the chromosomes at the INM, whereas KASH5 interacts with cytoplasmic motor proteins at the ONM, establishing a transmembrane linkage complex that can transduce cytoskeletal force to the telomeres, thus driving chromosomal movement (Hiraoka and Dernburg 2009; Starr and Fridolfsson 2010). Testis suspensions were stained for lamin B and TRF1 to evaluate telomeric function in spermatocytes from P06. The results demonstrated that most of the TRF1-labeled telomeres in spermatocytes from the control OA patient had been inserted into the NE, as identified by lamin B staining (Fig. 4A). Conversely, the telomeres in most spermatocytes from P06 remained within the nucleus (Fig. 4A, B), although the total amount and expression level of TRF1 did not change significantly (Fig. 4C, D). These results suggest that SUN1 is necessary for telomeric NE attachment in human spermatocytes.
SUN1 deficiency in spermatocytes causes the detachment of telomeres from the nuclear envelope. A Equatorial images of structurally preserved spermatocytes from P06 or OA patients stained using antibodies specific for the indicated proteins. B Quantification revealed that the numbers of internal TRF1 foci within spermatocytes from P06 were significantly increased relative to those in control samples. C The total numbers of TRF1 foci of the spermatocytes from P06 were comparable to the OA group. D Graphs showing the TRF1 intensities per cell were comparable between the two groups. Over 50 cells were analyzed per group for all analyses. TRF2 telomeric repeat-binding factor 1. Ns, P > 0.05; ****P < 0.0001
The SUN1–KASH5 complexes were almost absent in the SUN1-deficient spermatocytes
Due to the formation of the LINC complex with SUN1, the location of KASH5 in ONM is closely related to the location of SUN1 in INM (Wu et al. 2022). To evaluate the adverse effects of SUN1 loss on the KASH5, the expression of KASH5 was also analyzed in these spermatocytes. The immunofluorescence revealed a reduction of KASH5 at the ONM relative to control samples (Fig. 5A, B). Further, Western blot analysis showed that the KASH5 level was significantly decreased in the testes of P06 (Fig. 5C). The expression levels of TRF1 and TRF2 did not change significantly (Fig. 5C). These findings support that loss of SUN1 in human spermatocytes leads to significant abnormalities in KASH5 expression and localization. Moreover, the observed meiotic arrest in this NOA patient may have been caused by a severe disruption of the interactions between the telomere, SUN1, and KASH5.
KASH5 aberrations in the SUN1-deficient spermatocytes. A Representative images of surface-spread spermatocytes in control and P06 samples stained with antibodies specific for KASH5 (green) and lamin B (red), with the KASH5 signal on the NE marked with an arrow. B Quantifying the number of KASH5 loci in equatorial images of spermatocytes in P06 and OA samples. Over 50 cells were analyzed per group for all analyses. C Western blot revealed a reduced level of KASH5 in the SUN1-deficient testes. TRF1\2, telomeric repeat-binding factor 1\2. ****P < 0.0001
Discussion
Meiosis is a strictly regulated and highly conserved mechanism in eukaryotes to sustain genetic variety and the long-term survival of each species. When meiosis-related proteins exhibit congenital abnormalities that impact protein structural and/or functional characteristics, the results generally lead to detrimental outcomes and impaired gametogenesis, as in the case of mutations in the TEX11 (Yatsenko et al. 2015), MSH4 (Wyrwoll et al. 2021), MSH5 (Gong et al. 2022), and SHOC1 (Yao et al. 2021) genes. In this case, a Chinese patient with NOA from a consanguineous family was found to have a homozygous SUN1 variation [c. 663C > A:p.Tyr221X]. This mutation was related with the degradation of SUN1 mRNA and poor INM localization in this patient's spermatocytes. Previous studies have shown that KASH5 deficiencies are associated with azoospermia stemming from meiotic arrest due to poor telomere-SUN1–KASH5 interactions (Wu et al. 2022; Yang et al. 2022). Our results demonstrate that this novel biallelic SUN1 variant may contribute to the pathogenesis of NOA in humans by disrupting these interactions.
Coordinated mechanical force-mediated chromosomal movement is essential for homologous pairing, synapsis, and eventual segregation (Shibuya et al. 2015). Extensive research conducted in a range of species has shown that the force-transducing system, which coordinates the movement of chromosomes during prophase I, is highly conserved, consisting of cytoplasmic motor proteins responsible for mechanical force generation as well as the LINC complex that spans the NE and facilitates force transduction (Lee and Burke 2018). Moreover, the binding of telomeres to the NE and LINC complex is regulated by the TTM complex (Long et al. 2017; Shibuya et al. 2015). When any of the proteins involved in this process are absent, meiotic progression is disrupted, leading to meiotic arrest (Kmonickova et al. 2020). In patient P06 in this study, complete meiotic arrest of spermatogenesis was observed, and no spermatocytes were detected at the pachytene-like stage. This suggests that meiotic progression was likely dysregulated such that arrest occurred during the leptotene or zygotene stages, in line with results reported in Sun1-KO mice (Ding et al. 2007).
SUN1 and KASH domain proteins, connected to the INM and the ONM, respectively, make up the LINC complex (Shibuya et al. 2015). This SUN–KASH complex is considered a key mediator of meiotic telomere attachment and movement (Link et al. 2015), given that these functions are profoundly impaired in mammals and even budding yeast harboring mutations in the genes that encode components of this complex (Bupp et al. 2007; Ding et al. 2007; Horn et al. 2013; Schober et al. 2009). Prior murine research has demonstrated that SUN1 is a key LINC complex component responsible for controlling chromosomal movement during prophase I (Ding et al. 2007). While all mice lacking Sun1 expression are infertile, further studies have shown that male Sun1-KO mice exhibit arrested spermatogenesis at prophase I due to impaired telomere attachment to the cytoskeleton via the NE (Ding et al. 2007). Despite extensive study of the LINC complex in mice, its function in human fertility remains to be fully documented. Recent work has highlighted a link between certain KASH5 variants and NOA pathogenesis (Fakhro et al. 2018; Wu et al. 2022; Zhang et al. 2022). Like murine KASH5, human cells express KASH6 in several foci around the NE and at chromosome ends. When KASH5 is absent, it leads to a loss of SUN1, resulting in a loss of function of the LINC complex on NE, causing impaired chromatin telomeric attachment, aberrant synapsis, dysfunctional DSB repair, and meiotic arrest (Fakhro et al. 2018; Wu et al. 2022; Zhang et al. 2022). Here, telomere attachment was similarly impaired in spermatocytes from an NOA patient exhibiting SUN1 deficiency. In this patient, most telomeres in spermatocytes were restricted to the nucleus, and significant reductions in KASH5 NE localization were observed in these cells suggesting that the loss of SUN1 could make SUN1 lose function, leading to the disruption of telomere-SUN1–KASH5 interactions. In addition, unlike CCDC155, SUN1 is not a testicle-specific expressed protein. Our research also indicated that while patients lacking SUN1 have NOA, the rest of their bodies may be healthy. These results are comparable to an in vivo study in mice, which reported similar findings. (Ding et al. 2007).
Overall, these results highlight the conservation of SUN1 as a mediator of meiotic progression across species while offering insight into a new potential genetic driver of NOA pathogenesis.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Anderson J, Farr S, Jamieson D, Warner L, Macaluso MJF (2009) Infertility services reported by men in the United States: National Survey Data. Fertil Steril 91:2466–2470. https://doi.org/10.1016/j.fertnstert.2008.03.022
Arshad M, Majzoub A, Esteves SJ (2020) Predictors of surgical sperm retrieval in non-obstructive azoospermia: summary of current literature. Int Urol Nephrol 52:2015–2038. https://doi.org/10.1007/s11255-020-02529-4
Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD (2005) SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118:3233–3245. https://doi.org/10.1242/jcs.02466
Bolcun-Filas E, Handel MA (2018) Meiosis: the chromosomal foundation of reproduction. Biol Reprod 99:112–126. https://doi.org/10.1093/biolre/ioy021
Bupp JM, Martin AE, Stensrud ES, Jaspersen SL (2007) Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179:845–854. https://doi.org/10.1083/jcb.200706040
Burke B (2018) LINC complexes as regulators of meiosis. Curr Opin Cell Biol 52:22–29. https://doi.org/10.1016/j.ceb.2018.01.005
Chang YF, Imam JS, Wilkinson MF (2007) The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76:51–74. https://doi.org/10.1146/annurev.biochem.76.050106.093909
Chen Y, Wang Y, Chen J, Zuo W, Fan Y, Huang S, Liu Y, Chen G, Li Q, Li J, Wu J, Bian Q, Huang C, Lei M (2021) The SUN1-SPDYA interaction plays an essential role in meiosis prophase I. Nat Commun 12:3176. https://doi.org/10.1038/s41467-021-23550-w
Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M (2007) SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12:863–872. https://doi.org/10.1016/j.devcel.2007.03.018
Fakhro KA, Elbardisi H, Arafa M, Robay A, Rodriguez-Flores JL, Al-Shakaki A, Syed N, Mezey JG, Abi Khalil C, Malek JA, Al-Ansari A, Al Said S, Crystal RG (2018) Point-of-care whole-exome sequencing of idiopathic male infertility. Genet Med 20:1365–1373. https://doi.org/10.1038/gim.2018.10
Gershoni M, Hauser R, Barda S, Lehavi O, Arama E, Pietrokovski S, Kleiman SJ (2019) A new MEIOB mutation is a recurrent cause for azoospermia and testicular meiotic arrest. Hum Reprod 34:666–671. https://doi.org/10.1093/humrep/dez016
Gong C, Abbas T, Muhammad Z, Zhou J, Khan R, Ma H, Zhang H, Shi Q, Shi B (2022) A homozygous loss-of-function mutation in MSH5 abolishes MutSγ axial loading and causes meiotic arrest in NOA-affected individuals. Int J Mol Sci. https://doi.org/10.3390/ijms23126522
Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117:4025–4032. https://doi.org/10.1242/jcs.01363
Hiraoka Y, Dernburg AF (2009) The SUN rises on meiotic chromosome dynamics. Dev Cell 17:598–605. https://doi.org/10.1016/j.devcel.2009.10.014
Horn HF, Kim DI, Wright GD, Wong ES, Stewart CL, Burke B, Roux KJ (2013) A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol 202:1023–1039. https://doi.org/10.1083/jcb.201304004
Jarvi K, Lo K, Fischer A, Grantmyre J, Zini A, Chow V, Mak VJ (2010) CUA guideline: the workup of azoospermic males. Can Urol Assoc J 4:163–167. https://doi.org/10.5489/cuaj.10050
Kmonickova V, Frolikova M, Steger K, Komrskova K (2020) The role of the LINC complex in sperm development and function. Int J Mol Sci. https://doi.org/10.3390/ijms21239058
Krausz C, Riera-Escamilla A (2018) Genetics of male infertility. Nat Rev Urol 15:369–384. https://doi.org/10.1038/s41585-018-0003-3
Lee YL, Burke B (2018) LINC complexes and nuclear positioning. Semin Cell Dev Biol 82:67–76. https://doi.org/10.1016/j.semcdb.2017.11.008
Link J, Jahn D, Alsheimer M (2015) Structural and functional adaptations of the mammalian nuclear envelope to meet the meiotic requirements. Nucleus 6:93–101. https://doi.org/10.1080/19491034.2015.1004941
Long J, Huang C, Chen Y, Zhang Y, Shi S, Wu L, Liu Y, Liu C, Wu J, Lei M (2017) Telomeric TERB1-TRF1 interaction is crucial for male meiosis. Nat Struct Mol Biol 24:1073–1080. https://doi.org/10.1038/nsmb.3496
Majzoub A, Arafa M, Clemens H, Imperial J, Leisegang K, Khalafalla K, Agarwal A, Henkel R, Elbardisi H (2022) A systemic review and meta-analysis exploring the predictors of sperm retrieval in patients with non-obstructive azoospermia and chromosomal abnormalities. Andrologia 54:e14303. https://doi.org/10.1111/and.14303
Mikolcevic P, Isoda M, Shibuya H, del Barco BI, Igea A, Suja JA, Shackleton S, Watanabe Y, Nebreda AR (2016) Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat Commun 7:11084. https://doi.org/10.1038/ncomms11084
Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198:165–172. https://doi.org/10.1083/jcb.201204085
Mühlemann O (2016) Spermatogenesis studies reveal a distinct nonsense-mediated mRNA decay (NMD) mechanism for mRNAs with long 3’UTRs. PLoS Genet 12:e1005979. https://doi.org/10.1371/journal.pgen.1005979
Parvanov ED, Petkov PM, Paigen K (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327:835. https://doi.org/10.1126/science.1181495
Peters A, Plug A, van Vugt M, de Boer PJC (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5:66–68. https://doi.org/10.1023/a:1018445520117
Salas-Huetos A, Tüttelmann F, Wyrwoll MJ, Kliesch S, Lopes AM, Goncalves J, Boyden SE, Wöste M, Hotaling JM, Nagirnaja L, Conrad DF, Carrell DT, Aston KI (2021) Disruption of human meiotic telomere complex genes TERB1, TERB2 and MAJIN in men with non-obstructive azoospermia. Hum Genet 140:217–227. https://doi.org/10.1007/s00439-020-02236-1
Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2:621–627. https://doi.org/10.1038/35085086
Scherthan H (2007) Telomere attachment and clustering during meiosis. Cell Mol Life Sci 64:117–124. https://doi.org/10.1007/s00018-006-6463-2
Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134:1109–1125. https://doi.org/10.1083/jcb.134.5.1109
Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM (2009) Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23:928–938. https://doi.org/10.1101/gad.1787509
Shibuya H, Hernandez-Hernandez A, Morimoto A, Negishi L, Hoog C, Watanabe Y (2015) MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163:1252–1266. https://doi.org/10.1016/j.cell.2015.10.030
Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26:421–444. https://doi.org/10.1146/annurev-cellbio-100109-104037
Wu H, Zhang X, Hua R, Li Y, Cheng L, Li K, Liu Y, Gao Y, Shen Q, Wang G, Lv M, Xu Y, He X, Cao Y, Liu MJ (2022) Homozygous missense mutation in CCDC155 disrupts the transmembrane distribution of CCDC155 and SUN1, resulting in non-obstructive azoospermia and premature ovarian insufficiency in humans. Hum Genet. https://doi.org/10.1007/s00439-022-02459-4
Wyrwoll MJ, van Walree ES, Hamer G, Rotte N, Motazacker MM, Meijers-Heijboer H, Alders M, Meißner A, Kaminsky E, Wöste M, Krallmann C, Kliesch S, Hunt TJ, Clark AT, Silber S, Stallmeyer B, Friedrich C, van Pelt AMM, Mathijssen IB, Tüttelmann F (2021) Bi-allelic variants in DNA mismatch repair proteins MutS homolog MSH4 and MSH5 cause infertility in both sexes. Hum Reprod 37:178–189. https://doi.org/10.1093/humrep/deab230
Xie C, Wang W, Tu C, Meng L, Lu G, Lin G, Lu LY, Tan YQ (2022) Meiotic recombination: insights into its mechanisms and its role in human reproduction with a special focus on non-obstructive azoospermia. Hum Reprod Update. https://doi.org/10.1093/humupd/dmac024
Yang C, Lin X, Ji Z, Huang Y, Zhang L, Luo J, Chen H, Li P, Tian R, Zhi E, Hong Y, Zhou Z, Zhang F, Li Z, Yao C (2022) Novel bi-allelic variants in KASH5 are associated with meiotic arrest and non-obstructive azoospermia. Mol Hum Reprod. https://doi.org/10.1093/molehr/gaac021
Yao C, Yang C, Zhao L, Li P, Tian R, Chen H, Guo Y, Huang Y, Zhi E, Zhai J, Sun H, Zhang J, Hong Y, Zhang L, Ji Z, Zhang F, Zhou Z, Li Z (2021) Bi-allelic SHOC1 loss-of-function mutations cause meiotic arrest and non-obstructive azoospermia. J Med Genet 58:679–686. https://doi.org/10.1136/jmedgenet-2020-107042
Yatsenko A, Georgiadis A, Röpke A, Berman A, Jaffe T, Olszewska M, Westernströer B, Sanfilippo J, Kurpisz M, Rajkovic A, Yatsenko S, Kliesch S, Schlatt S, Tüttelmann FJT (2015) X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372:2097–2107. https://doi.org/10.1056/NEJMoa1406192
Yu X, Li M, Cai F, Yang S, Liu H, Zhang HJ (2021) TEX11A new mutation causes azoospermia and testicular meiotic arrest. Asian J Androl 23:510–515. https://doi.org/10.4103/aja.aja_8_21
Zhang Q, Tao C, Gao S, Li S, Xu B, Ke H, Wang Y, Zhang F, Qin Y, Zhang L, Guo T (2022) Homozygous variant in KASH5 causes premature ovarian insufficiency by disordered meiotic homologous pairing. J Clin Endocrinol Metab. https://doi.org/10.1210/clinem/dgac368
Zheng B, Zhao D, Zhang P, Shen C, Guo Y, Zhou T, Guo X, Zhou Z, Sha J (2015) Quantitative proteomics reveals the essential roles of stromal interaction molecule 1 (STIM1) in the testicular cord formation in mouse testis. Mol Cell Proteomics 14:2682–2691. https://doi.org/10.1074/mcp.M115.049569
Acknowledgements
We would like to thank the families that participated and supported this study.
Funding
This work was supported by the National Natural Science Foundation of China (82201762 and 82201765), the Natural Science Foundation of Jiangsu Province (BK20220200), the Suzhou Science and Technology Development Plan (LCZX202109), the Science and Technology Project of Changzhou (CJ20220143), and the Science and Technology Project of Jiangsu Health Committee (Z2021048).
Author information
Authors and Affiliations
Contributions
Conceptualization: TG, QZ, and HL. Methodology QM, BS, and DZ. Validation: QM, BS, and DZ. Formal analysis: QM, BS, and DZ. Resources: XF and JW. Data curation: JW. Writing—original draft preparation: QM. Writing—review and editing: TG, QZ, and HL. Visualization: XF. Supervision: HL. Project administration: QZ. Funding acquisition: QM, DZ, QZ, TG. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
The study was conducted per the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Suzhou Municipal Hospital, China (No. 2020190).
Consent to participate
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from the patient(s) to publish this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meng, Q., Shao, B., Zhao, D. et al. Loss of SUN1 function in spermatocytes disrupts the attachment of telomeres to the nuclear envelope and contributes to non-obstructive azoospermia in humans. Hum Genet 142, 531–541 (2023). https://doi.org/10.1007/s00439-022-02515-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-022-02515-z