Abstract
It is not well known whether genetic markers identified through genome-wide association studies (GWAS) confer similar or different risks across people of different ancestry. We screened a regularly updated catalog of all published GWAS curated at the NHGRI website for GWAS-identified associations that had reached genome-wide significance (p ≤ 5 × 10−8) in at least one major ancestry group (European, Asian, African) and for which replication data were available for comparison in at least two different major ancestry groups. These groups were compared for the correlation between and differences in risk allele frequencies and genetic effects’ estimates. Data on 108 eligible GWAS-identified associations with a total of 900 datasets (European, n = 624; Asian, n = 217; African, n = 60) were analyzed. Risk-allele frequencies were modestly correlated between ancestry groups, with >10% absolute differences in 75–89% of the three pairwise comparisons of ancestry groups. Genetic effect (odds ratio) point estimates between ancestry groups correlated modestly (pairwise comparisons’ correlation coefficients: 0.20–0.33) and point estimates of risks were opposite in direction or differed more than twofold in 57%, 79%, and 89% of the European versus Asian, European versus African, and Asian versus African comparisons, respectively. The modest correlations, differing risk estimates, and considerable between-association heterogeneity suggest that differential ancestral effects can be anticipated and genomic risk markers may need separate further evaluation in different ancestry groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Technological advances have greatly increased the availability and reduced the cost of genetic information (Ashleyet al. 2010; Janssens et al. 2008). Genome-wide association studies (GWAS) addressing a variety of common human diseases have found many hundreds of associations with robust statistical support heightening the expectations for a substantial contribution of genomics to personalized medicine (Hindorff et al. 2009, 2011; McCarthy et al. 2008). Most GWAS have been performed on European descent populations, but increasing numbers of such investigations are now performed on other ancestral groups. In the currently available GWAS literature, there are occasions where the observed genetic effects seem to exhibit effect consistency across ancestral groups, either in terms of effect direction or comparability of the magnitude of the effect (Waters et al. 2010). On the other hand, there are other occasions where GWAS-derived signals point to a pattern of differential ancestral effects (Rosenberg et al. 2010).
Identifying patterns of observed genetic effect variability or consistency across populations of differing ancestry (Ioannidis et al. 2004) can support our understanding of the genetic architecture of complex diseases. The risk conferred by GWAS-identified markers may vary in people of different ancestry (Rosenberg et al. 2010; Helgason et al. 2007) due to variability in allelic frequencies and to differences in linkage disequilibrium (LD) between the identified variants and the true functional variants that underlie disease risk. Alternatively, if common variant associations are consistent across major ethnic groups, the causal variants they presumably tag are also likely to be common, arguing against the synthetic association model in those instances (Waters et al. 2010). It is also possible that the true functional variants may not be the same in different ancestry groups (Ioannidis et al. 2004; Manica et al. 2005; Tang 2006). An unequal distribution of disease-associated alleles between different ancestry populations has been described for several recessive mendelian disorders, e.g., hemochromatosis, and for some complex disorders (e.g., inflammatory bowel disease or cardiomyopathies) (Burchard et al. 2003; Dhandapany et al. 2009). Typical reasons for this are population-specific mutations, different LD patterns and recombination events, or even differing selective pressures in the areas of origin or residence of these groups. Furthermore, individuals of European and African ancestry differ significantly in the expression of many genes which could contribute to some of the observed differences in susceptibility to common diseases (Spielman et al. 2007; Zhang et al. 2008). Finally, differences in risk-allele frequencies may affect the power to detect genome-wide significant associations across populations of varying ancestral origin (Moonesinghe et al. 2008), and may also affect the transferability of disease-risk prediction across major populations. Such differences might require consideration of ancestral origin to control for unmeasured allele frequency differences as well as measured allele frequencies and LD differences in developing personalized prediction models.
Empirical assessment of consistency of genetic effects for GWAS-identified markers in populations of variable ancestral origin can thus shed light on the genetic architecture of common diseases as well as potential for generalizability of findings across population groups. Here, we evaluated whether frequencies of the genetic markers of interest and the genetic effects that they confer are different across populations of different ancestral origin for 108 GWAS-discovered markers with robust statistical support.
Methods
Eligible associations
We evaluated GWAS-discovered associations for any phenotype or trait that (a) have had robust statistical support in at least one major ancestral group and (b) had been finally replicated in at least two of the three different major ancestral groups (see below for ancestral origin and final replication definitions).
Ancestral origin
We used the following categorization of self-reported ancestral origin that is in accordance with a previous empirical evaluation of ancestry differences for candidate gene associations (Ioannidis et al. 2004). ‘European ancestry’ was assigned to native populations of Europe and to people of European origin living in Oceania, North America and South America, excluding Hispanics (Spanish speaking people living in the Americas). As ‘African ancestry’, we considered populations native to or with origin in sub-Saharan Africa, and self-identified African Americans. ‘Asian ancestry’ was assigned to native populations of China, Japan, Korea, and Taiwan, excluding India, Indochina, and Philippines. In HapMap terms, CEU, YRI, and CHB + JPT panels would belong to our European, African, and Asian groups, respectively (The International HapMap Project 2003). We excluded upfront populations not included in the groups above.
Robust statistical support
Robust statistical support was defined as having p ≤ 5 × 10−8 (genome-wide significance, GWS) in a meta-analysis combining with fixed effects all available datasets from a specific ancestry group in at least one genome-wide association study publication. We included both agnostic discovery and replication data in the calculations for attaining GWS.
Screened GWAS publications and selection of associations and datasets
We screened a regularly updated list of all GWAS publications curated at the NHGRI website (Hindorff et al. 2011) through January 19, 2010 when 2,659 associations at p values ≤10–5 had been entered in the database. The NHGRI database includes GWAS that have attempted to genotype 100,000 SNPs or more. Full-text articles were obtained and further scrutinized, including all the supplements of each publication.
We considered GWAS that evaluated more than one of the previously defined ancestry groups at a final replication stage in the same publication; or compared their data against previously published data from one or more GWAS that had investigated the same phenotype in populations of different ancestry (see Supplementary Methods). Different associations were defined by differences in gene variant, phenotype, or both, but not just by different genetic models of inheritance (e.g., allele-based vs. genotype-based); whenever highly linked markers were detected (r 2 > 0.8), we kept the one with the lowest p value. Further details on selection of associations appear in Supplementary Methods. Most GWAS use data from diverse pre-existing studies in the discovery and/or replication phase. We a priori endorsed the definition of distinct population datasets as reported in the original papers. To avoid the inflation of effect sizes due to the winner’s curse (Ioannidis 2007; Zollner and Pritchard 2007), we only selected datasets that pertained to the final replication stage.
Statistical analysis
Risk-allele frequencies
For each eligible association, we recorded the frequencies of the risk allele in the European (CEU), African (YRI), and Asian (CHB + JPT) descent populations in HapMap phase 2 data release 27, February 2009 (The International HapMap 2003), on NCBI B36 assembly, dbSNP b126. We estimated the Pearson correlation coefficient between the risk-allele frequencies in the CEU population and the respective frequencies in the CHB + JPT or YRI populations. We used an inverse-variance weighted summary estimate for the combined Asian datasets (CHB + JPT) after an arcsin transformation. Less than a third of the GWAS provided genotype counts in each dataset and group to allow using the risk-allele frequency information from these studies, but data reported were generally compatible with their respective HapMap estimates. To evaluate population differentiation, we also extracted information on F st estimates for all assessed genetic variants as provided at HapPlotter (Voight et al. 2006).
Genetic effect sizes
We used the odds ratio (OR) for binary outcomes and the standardized mean difference (SMD) for continuous outcomes. For consistency in the analyses, genetic effect estimates were always presented so as to have OR > 1 or SMD > 0 in the ancestry group reaching overall GWS. We synthesized effect sizes (OR or SMD) for datasets in each association separately for each ancestry group (Supplementary Methods). Many GWAS did not provide effect size information per dataset, and had already combined data under fixed effects models. Therefore, for consistency we also combined all remaining data with fixed effects models (Mantel and Haensel 1959; Cooper and Hedges 1994, DerSimonian and Laird 1986). Fixed effects models have better power for discovery, but random effects are more appropriate for estimating the typical effects and their expected variability across different populations (Pereira et al. 2009).
We evaluated how often effect sizes differed more than twofold between the compared ancestry groups. This includes cases where effect estimates were in the opposite direction (e.g., OR 1.20 in Europeans and 0.90 in Asians), and cases where effect estimates were in the same direction, but the OR-1 was more than double in one ancestry group than the other (e.g., OR 1.20 in Europeans and <1.10 in Asians). We also evaluated whether the effect sizes differed beyond chance in populations of different ancestry, using the Z score as described previously (Cappelleri et al. 1996; Ioannidis et al. 2001). Since point estimates alone do not account for the uncertainty surrounding them and testing effect sizes for significant differences depends on whether there is limited (underpowered) or extensive (overpowered) evidence for the compared ancestry groups, these analyses offer complementary information. We also estimated the Pearson correlation coefficient between the effect sizes for a specific association in European ancestry populations and the respective effect sizes in Asian or African ancestry populations. This was done separately for ORs (n = 68) and for SMDs (n = 40). Combined analyses translated SMDs to OR equivalent using the conversion factor 1.81, as previously proposed (Chinn 2000). The conversion (1.81 = π/√3) works well for normally distributed effects (as assumed in OR and SMD calculations in general). We also sought to explore whether very small effect sizes could dilute the observed correlation estimates due to simple stochastic variation or whether associations involving uncommon and rare variants could exhibit a different pattern of correlation due to their differing evolutionary characteristics. We thus performed a sensitivity analysis limited to associations where the effect size in the ancestry group that reached GWS corresponded to an OR > 1.2 and another sensitivity analysis limited to associations where the assessed variants were rare (maf ≤ 0.01) or uncommon (maf ≤ 0.05) in at least one of the compared ancestral groups as estimated in the HapMap phase 2 data.
Additionally, for each association we calculated the pairwise relative odds ratio (ROR) for every pair of ancestry groups, by dividing the summary ORs or the OR-translated SMDs of the corresponding ancestry groups. For example, an association with a European–Asian ROR > 1 means that the genetic effect estimate observed in the combined European datasets was larger compared to the effect estimate observed in datasets of Asian ancestry. We then calculated the summary ROR (sROR) across all associations combining the natural logarithms of all individual RORs for each ancestral between-group comparison, using a random effects model to allow for the large heterogeneity in ROR estimates and then re-exponentiating the derived summary log ROR. Heterogeneity in the ROR estimates was estimated with the Q test (considered significant for p < 0.10) and the I 2 metric and its 95% CI (Higgins et al. 2003; Ioannidis et al. 2007).
Adjacent hotspots
Since the observed differences in the genetic risk estimates across ancestral groups could be explained by differences in linkage disequilibrium (LD) between the identified variants and the true functional variants that underlie disease risk, we looked for the presence of recombination hotspots in vicinity to the GWAS-discovered genetic markers. For every assessed SNP, the current mapping position was retrieved through the 1000 Genomes browser (1000 Genomes 2011). We then identified 1000 Genomes hotspots within a 200-kb distance around the SNP under study and captured the number of nearby hotspots as well as the distance from the SNP under study to the closest end of the most adjacent hotspot. Univariate logistic regression analyses were further performed to evaluate whether the numbers of adjacent hotspots or the distance to the nearest hotspot could predict the observed ROR estimates or the occurrence of risk estimate differences (statistically significantly different ancestral estimates, opposite direction genetic ancestral effects or more than twofold differences across ancestries). For the risk estimate differences’ logistic regression analyses, separate analyses were performed to predict the occurrence of differences in all pairwise comparisons and the occurrence of any kind of risk estimate difference across all ancestral groups for every assessed association.
Analyses were performed in Stata SE 10.0 (College Station, TX, USA). All p values are two tailed.
Results
Characteristics of the eligible associations
We screened 365 GWAS reports published through January 19, 2010. Of those, we considered eligible for our analysis, all the GWAS literature on associations that had been assessed in at least two of the three major ancestry groups (Europeans, Asians, Africans) with GWS in at least one ancestry group. Forty-one publications qualified for inclusion in our study and they assessed 151 potentially eligible associations. Eighteen associations pertaining to 13 publications were further excluded due to lack of available data for analysis (effect size estimates and standard errors or p values) and 4 associations were also excluded because meta-analysis per ancestry group did not reach GWS in any ancestry group. Finally, 21 associations were excluded due to the presence of another highly linked marker with a lower p value.
The 108 eligible associations of 105 SNPs with various outcomes were published in 33 papers [Table 1, Appendix A (Supplementary Methods)]. Binary disease outcomes were assessed in 68 (63%) associations and 40 associations assessed continuous traits (mostly height [n = 29], but also body mass index, eosinophil count, and uric acid). After excluding discovery and trimming down datasets (see Supplementary Methods for definitions), there were 900 eligible final replication datasets. These datasets, where variants surviving the previous stage(s) were assessed without other subsequent validation, would be least likely to be affected by the “winner’s curse” phenomenon (Campbell et al. 2005; Yang et al. 2008; Tang et al. 2005) that tends to inflate the estimates of newly discovered associations passing a given discovery threshold of significance. The median number of analyzed final replication datasets per association tested was 6 (range 2–27) and the median number of participants per association in these datasets was 23,321 (IQR 4,400–52,886) for all three ancestry groups combined.
Non-European populations were under-represented among the populations assessed in the 108 GWS associations. All eligible associations had been evaluated in populations of European ancestry (including 68 associations evaluated in a non-European study and contrasted with a previously published European genome-wide association study). A total of 97 associations were evaluated in both European and Asian populations; 24 in both European and African populations; and 13 in both Asian and African populations (these 13 associations had been evaluated in all three groups). Likewise, most final replication datasets included individuals of European ethnic origin (n = 624), while populations of Asian or African ethnic origin were represented by only 217 and 60 datasets, respectively. Log-additive models were reported in 35 (32%) associations, while in the rest 73 (72%) associations the effect size was calculated under an additive model assumption.
Risk-allele frequencies
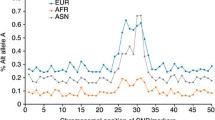
As shown in Figure and Supplementary Table 1, risk-allele frequencies had a modest correlation between different ancestry groups. CHB and JPT minor allele frequencies were highly correlated, as expected (correlation coefficient = 0.98, p < 0.001), and therefore merged into a combined CHB + JPT group. The Pearson correlation coefficient was 0.63 (p < 0.001) for frequencies in the CEU versus CHB + JPT, 0.53 (p < 0.001) for CEU versus YRI, and 0.31 (p = 0.01) for CHB + JPT versus YRI ancestry. Differences in the minor allele frequencies of >10% in absolute frequency were common and occurred in 77 (72%), 75 (69%), and 89 (83%) of the CEU versus CHB + JPT, CEU versus YRI and CHB + JPT versus YRI comparisons. Most of the variants were common in at least one ancestral group (minor allele frequency >5%). According to HapMap data, 7, 20, and 15 genetic variants were uncommon in CEU, CHB + JPT and YRI samples, respectively, while 2, 9 and 8 rare variants were observed in the aforementioned ancestral groups, respectively. The risk allele was the minor allele in 49, 60, and 54 of the assessed associations in CEU, CHB + JPT, and YRI samples, respectively. Of the assessed genetic variants, 8 and 8 had F st > 0.25 for the CEU populations compared to the CHB + JPT and YRI samples, respectively. These were variants associated with breast cancer, schizophrenia, height, uric acid, systemic lupus erythematosus, Parkinson’s disease (Supplementary Table 2).
Genetic effect sizes
As shown in Table 2 effect sizes showed notable variability across different ancestry groups, particularly for comparisons with African ancestry groups. Among the European and Asian ancestry genetic effect size estimates, 17 (18%) had estimates in the opposite direction and another 31 (39%) were in the same direction, but differed from more than twofold; effect size estimates differed beyond chance in 21 of the 97 (22%) comparisons. The frequency of discrepancies between European and African effect size estimates was 5 (21%), 11 (58%) and 10 (42%), respectively. The frequency of discrepancies between Asian and African effect size estimates was 5 (39%), 4 (50%) and 3 (23%), respectively.
Regarding the potential effect of the underlying LD on the observed effect concordance patterns, there were 7 SNPs with no recombination hotspot nearby. Overall, a median of 2 (range 0–6) hotspots were found in vicinity to the assessed SNPs and the median distance to the closest hotspot was 23.8 kb; neither the number of identified hotspots nor the distance to the closest hotspot seem to be a statistically significant predictor of finding risk estimate differences in any of the three aspects studied (statistically significant differences; opposite direction of effects; opposite direction of effects or same direction but with more than twofold difference). In a pairwise fashion, the number of nearby hotspots had a nominally statistically significant inverse weak relationship with the occurrence of more than twofold genetic risk differences (OR 0.70; 95% CI 0.51–0.97; p = 0.031) for the European–Asian comparison, but this should be interpreted cautiously given the number of analyses (Table 3).
Overall, when all 108 GWS associations were considered, there was moderate correlation among the effect sizes in different ancestry groups (Fig. 1): the correlation coefficients were 0.33 (p < 0.001), 0.27 (p = 0.20) and 0.20 (p = 0.51) for the European–Asian, European–African and Asian–African comparisons. When limited to the associations where the effect size in the ancestry group that reached GWS corresponded to an OR > 1.2, the correlation coefficient was 0.19 (p = 0.23) for the European–Asian (n = 44) comparison (data were too limited for the other two comparisons). Although the assessed GWAS were designed to cover genetic signals corresponding to common variants, uncommon and even rare variants are represented in the GWAS genotyping platforms, albeit unevenly. The number of available rare variants did not allow for further analysis and, when limited to associations where the variant under study was uncommon in at least one of the compared ancestral groups, the correlation coefficient was 0.29 (p = 0.17) for the European–Asian (n = 24) comparison (data were too limited for the other two comparisons).
The ratio of the genetic effect (odds ratio) in one ancestry group versus another gives the ROR for an association. The ROR estimates can then be evaluated in a meta-analysis across all associations for each ancestry group contrast. As shown in Supplementary Figure, in many associations, the European estimates were much larger than the Asian estimates, and in an almost equal number of associations, the reverse was the case. Overall the summary ROR was very close to 1.00 (random-effects ROR = 1.08; 95% CI 1.03, 1.13) when all binary associations were considered, indicating that on average the genetic risk estimates did not differ between European and Asian groups; however, the observed between-association heterogeneity was large (I 2 = 75%) meaning that the observed variation in the ROR estimates across the three groups cannot be explained by chance alone. On average, African populations tended more frequently to have lower estimates of genetic risk compared to European estimates, but again there was very large heterogeneity (summary random-effects ROR = 1.17; 95% CI 1.06, 1.30; I 2 = 79%). The summary random-effects ROR was 1.03 (95% CI 0.92, 1.17; I 2 = 53%) for Asian versus African estimates. For the continuous traits, the summary random-effects ROR was 1.01 (95% CI 0.98, 1.04; I 2 = 63%) for the European-Asian comparisons and 1.06 (95% CI 0.96, 1.16; I 2 = 71% for the European–African comparisons). Considering binary and continuous associations together, the results did not change substantially (summary random-effects ROR = 1.04; 95% confidence interval: 1.02, 1.07; I 2 = 71% for the European–Asian comparisons, 1.13; 95% CI 1.05, 1.21; I 2 = 77% for the European–African comparisons and 1.01; 95% CI 0.90, 1.14; I 2 = 69% % for the Asian–African comparisons. Neither the number of nearby hotspots nor the distance to the closest hotspot was statistically significant associated with the observed ROR estimates for the assessed associations considering either pairwise or overall comparisons.
We next sought to explore whether the synthesis of data across all available ancestral groups would lead to enhancement of statistical significance of the replicated associations. Data synthesis under a fixed-effect model assumption across the assessed ancestral groups is shown in Supplementary Table 3. In 76 cases, the combined ancestry data yield a more promising (i.e., lower) p value while in 32 cases they yield a worse (i.e., higher) p value that is achieved by a single-ancestry analysis. Finally, among the 47 associations with available detailed information to estimate measures of between- and within-ancestry heterogeneity (τ2), the available data suggest more commonly larger variance compared to between-ancestry groups than within-ancestry groups, but these estimates have large uncertainty and for the majority of these associations there is not enough detailed information to calculate them (Supplementary Table 4).
Discussion
The current evaluation of 108 GWS associations with agnostically discovered genetic markers shows varying consistency in genetic effect sizes across major ancestral groups, and notable differences are encountered in a sizable proportion. Although the effect sizes seen in one population have modest correlation with the effect sizes in populations of another ancestry and it is not possible to reliably predict the effect in different ancestry groups for an association that has reached GWS in one ancestry group, effects differing beyond chance are less common. Combination of data from diverse ancestry groups may thus be more likely to lead to lower p values for association than ancestry-specific analyses.
A previous evaluation (Ioannidis et al. 2004) of 43 validated candidate gene associations had shown large differences in the allele frequencies, but quite good agreement with the effect sizes. While candidate gene associations tackled mostly variants that were thought to be the functional, causative variants, agnostic GWAS have captured common markers that are likely to be only in linkage disequilibrium with the culprits, and rarely the functional, causative variants themselves. The GWAS approach is far more efficient and has dramatically increased the yield of markers with robust support for association. Linkage disequilibrium of the discovered tagging markers with functional, causative variants may vary a lot across different ancestry groups (Bodmer and Bonilla 2008) affecting the correlations among observed cross-ancestry genetic-risk estimates. A previous study of population differentiation of GWAS-discovered SNPs for 26 conditions across different HapMap populations (Adeyemo and Rotimi 2010) found substantial differences in allele frequencies, but population differentiation (expressed by Fst) varied across different conditions. No previous study has examined differences in the genetic effects of GWAS-discovered variants across a large number of conditions. Several studies have tested a number of GWAS-discovered SNPs for one condition in different ancestry groups (Grant et al. 2008; Ioannidis 2009a, b; Ioannidis et al. 2009; Li et al. 2008; Ng et al. 2008; Yamada et al. 2009). Most have documented some modest or large differences in the genetic effects, but inferences are difficult to generalize from single studies and traits.
Some limitations should be discussed. First, although under adequate quality control procedures, genotyping is generally considered accurate (Chanock et al. 2007; Wellcome Trust Case Control Consortium 2007) in the GWAS era, other sources of errors, e.g., phenotype misclassification, or suboptimal characterization of ancestry groups could cause differences in different studies. Second, the notion that common ancestry is an efficient way to ensure population homogeneity has been extensively debated and ancestry definitions range from self-reported ancestry to reported grandparental birthplace to genome-defined (through hierarchical clustering methodology) ancestry; all approaches have limitations (Campbell et al. 2005; Yang et al. 2008; Tang et al. 2005; Royal et al. 2010). In all, genome-wide data are an accurate and cost-effective way to ascertain stratification within study populations, including stratification due to finer grained population histories largely unknown to individuals (Need and Goldstein 2006; Tian et al. 2008; Tishkoff et al. 2009). Conversely, genome-based clustering is not commonly used in replication datasets where ancestry is often assigned by self-report without genomic data confirmation. Third, comparison of genetic effects across ancestry groups may be influenced by the winner’s curse. We followed a strict protocol in selecting only final replication datasets (rather than initial discovery studies) that are least likely to be affected by the winner’s curse. Finally, we took extra care in assuring that the observed differences were observed in situations where there was strong evidence for the presence of an overall genetic association. We thus specifically focused on robustly replicated GWAS-derived associations, since we wanted to exclude the possibility of including null underlying associations where either the detection of any cross-ancestry difference would be due to chance alone or an observed pattern of consistency would reflect the null effect variation. Toward the same end and aiming to include associations less prone to selection and reporting biases, we excluded candidate gene studies and fully endorsed the agnostic, genome-wide association study framework.
On average the genetic effects were substantially smaller in African populations, probably because these 108 associations were generally not initially discovered in African populations. Of note, the majority of the assessed African populations were African–American groups where the proportion of European ancestry is considerable (as high as 20%) with very large variation among individuals (Bryc et al. 2010). Thus, the differences between European and African populations, other than African Americans, might be even more prominent.
The notion that the underlying recombination background could create between-ancestry and within-ancestry effect differences was not supported by our evaluation of recombination hotspots using 1000 Genomes data. Nevertheless, we should acknowledge that this analysis is not conclusive and it is limited due to the paucity of non-European population data in GWAS. Moreover, linkage disequilibrium patterns may differ enough to cause differences in associations between ancestral groups even when recombination hotspots are not identified in the 1000 Genomes data (Myers et al. 2005). Finally, as mentioned before for many associations, the amount of data from non-European populations was limited. Therefore, the proportion of the associations where there is a difference beyond chance in effect size across ancestry groups is probably underestimated.
The variable consistency in associations of common variants across major ancestry groups has important implications for understanding the genetic architecture of complex diseases. These differences may also reflect differences in the causal variants and/or their frequency across different populations. Inclusion of populations of differing ancestry in ongoing efforts should be further encouraged. The power to detect some markers may be different in one ancestry group than another, because of differences in allele frequencies, genetic effects, and potential environmental modifiers. Given that many loci may be pertinent to more than one ancestry group, one can obtain complementary lists of interesting loci with GWS signals by examining different ancestry groups, while an appropriately adjusted combined analysis will often increase power and contribute further discoveries. Finally, results of risk models involving many SNPs are more likely to be population-specific due to differences in LD patterns and allele frequencies characteristic of each composite SNP (Ransohoff and Khoury 2010; Yang et al. 2009), and should have separate validation in other populations. This has implications for the translational potential and development of genetic-risk prediction tests (Gulcher and Stefansson 2010; Ioannidis 2009a, b).
Abbreviations
- CEU:
-
Utah residents with Northern and Western European ancestry from the CEPH collection
- CHB:
-
Han Chinese in Beijing
- CI:
-
Confidence interval
- GWAS:
-
Genome-wide association study
- GWS:
-
Genome-wide significance
- IQR:
-
Inter-quartile range
- JPT:
-
Japanese in Tokyo
- LD:
-
Linkage disequilibrium
- NCBI:
-
National Centre for Biotechnology Information
- NHGRI:
-
National Human Genome Research Institute
- OR:
-
Odds ratio
- PMID:
-
PubMed identification number
- ROR:
-
Relative odds ratio
- SMD:
-
Standardized mean difference
- SNP:
-
Single nucleotide polymorphism
- YRI:
-
Yoruba in Ibadan
References
Adeyemo A, Rotimi C (2010) Genetic variants associated with complex human diseases show wide variation across multiple populations. Public Health Genomics 13:72–79
Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA et al (2010) Clinical assessment incorporating a personal genome. Lancet 375:1525–1535
Bodmer W, Bonilla C (2008) Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet 40:695–701
Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA et al (2010) Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA 107:786–791
Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Pérez-Stable EJ, Sheppard D, Risch N (2003) The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med 348:1170–1175
Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN (2005) Demonstrating stratification in a European American population. Nat Genet 37:868–872
Cappelleri JC, Ioannidis JP, Schmid CH, de Ferranti SD, Aubert M, Chalmers TC, Lau J (1996) Large trials vs meta-analysis of smaller trials: how do their results compare? JAMA 276:1332–1338
Chinn S (2000) A simple method for converting an odds ratio to effect size for use in meta-analysis. Stat Med 19:3127–3131
Cooper H, Hedges LV (eds) (1994) The handbook of research synthesis (Russell Sage 58. Foundation, New York)
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Tr 7:177–188
Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A et al (2009) A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet 41:187–191
1000 Genomes (2011) A deep catalog of human genetic variation. http://www.1000genomes.org/
Grant SF, Li M, Bradfield JP, Kim CE, Annaiah K, Santa E, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG et al (2008) Association analysis of the FTO gene with obesity in children of Caucasian and African ancestry reveals a common tagging SNP. PLoS One 3:e1746
Gulcher J, Stefansson K (2010) Genetic risk information for common diseases may indeed be already useful for prevention and early detection. Eur J Clin Invest 40:56–63
Helgason A, Pálsson S, Thorleifsson G, Grant SF, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I et al (2007) Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39:218–225
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106:9362–9367
Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA (2011) A catalog of published genome-wide association studies. (Available at: http://www.genome.gov/gwastudies)
Ioannidis JP (2007) Non-replication and inconsistency in the genome-wide association setting. Hum Hered 64:203–213
Ioannidis JP (2009a) Population-wide generalizability of genome-wide discovered associations. J Natl Cancer Inst 101:1297–1299
Ioannidis JP (2009b) Personalized genetic prediction: too limited, too expensive, or too soon? Ann. Intern Med 150:139–141
Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG (2001) Replication validity of genetic association studies. Nat Genet 29:306–309
Ioannidis JP, Ntzani EE, Trikalinos TA (2004) ‘Racial’ differences in genetic effects for complex diseases. Nat Genet 36:1312–1318
Ioannidis JP, Patsopoulos NA, Evangelou E (2007) Uncertainty in heterogeneity estimates in meta-analyses. BMJ 335:914–916
Ioannidis JP, Thomas G, Daly MJ (2009) Validating, augmenting and refining genome-wide association signals. Nat Rev Genet 10:318–329
Janssens AC, van Duijn CM (2008) Genome-based prediction of common diseases: advances and prospects. Hum Mol Genet 17:R166–R173
Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, Yu Z, Lin X (2008) Variants in the fat mass- and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes 57:264–268
Manica A, Prugnolle F, Balloux F (2005) Geography is a better determinant of human genetic differentiation than ethnicity. Hum Genet 118:366–371
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9:356–369
Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP (2008) Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci USA 105:617–622
Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310:321–324
NCI-NHGRI Working Group on Replication in Association Studies, Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D et al (2007) Replicating genotype-phenotype associations. Nature 447:655–660
Need AC, Goldstein DB (2006) Genome-wide tagging for everyone. Nat Genet 38:1227–1228
Ng MC, Park KS, Oh B, Tam CH, Cho YM, Shin HD, Lam VK, Ma RC, So WY, Cho YS et al (2008) Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and FTO in type 2 diabetes and obesity in 6, 719 Asians. Diabetes 57:2226–2233
Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JP (2009) Discovery properties of genome-wide association signals from cumulatively combined data sets. Am J Epidemiol 170:1197–1206
Ransohoff DF, Khoury MJ (2010) Personal genomics: information can be harmful. Eur J Clin Invest 40:64–68
Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M (2010) Genome-wide association studies in diverse populations. Nat Rev Genet 11:356–366
Royal CD, Novembre J, Fullerton SM, Goldstein DB, Long JC, Bamshad MJ, Clark AG (2010) Inferring genetic ancestry: opportunities, challenges, and implications. Am J Hum Genet 86:661–673
Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG (2007) Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet 39:226–231
Tang H (2006) Confronting ethnicity-specific disease risk. Nat Genet 38:13–15
Tang H, Quertermous T, Rodriguez B, Kardia SL, Zhu X, Brown A, Pankow JS, Province MA, Hunt SC, Boerwinkle E et al (2005) Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet 76:268–275
The International HapMap Project (2003) Nature 426:789–796
Tian C, Gregersen PK, Seldin MF (2008) Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet 17:R143–R150
Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O et al (2009) The genetic structure and history of Africans and African Americans. Science 324:1035–1044
Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72
Waters KM, Stram DO, Hassanein MT, Le Marchand L, Wilkens LR, Maskarinec G, Monroe KR, Kolonel LN, Altshuler D, Henderson BE et al (2010) Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet 6:e1001078
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678
Yamada H, Penney KL, Takahashi H, Katoh T, Yamano Y, Yamakado M, Kimura T, Kuruma H, Kamata Y, Egawa S et al (2009) Replication of prostate cancer risk loci in a Japanese case-control association study. J Natl Cancer Inst 101:1330–1336
Yang JJ, Burchard EG, Choudhry S, Johnson CC, Ownby DR, Favro D, Chen J, Akana M, Ha C, Kwok PY et al (2008) Differences in allergic sensitization by self-reported race and genetic ancestry. J Allergy Clin Immunol 122:820–827
Yang Q, Flanders WD, Moonesinghe R, Ioannidis JP, Guessous I, Khoury MJ (2009) Using lifetime risk estimates in personal genomic profiles: estimation of uncertainty. Am J Hum Genet 85:786–800
Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ et al (2008) Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet 82:631–640
Zollner S, Pritchard JK (2007) Overcoming the winner’s curse: estimating penetrance parameters from case–control data. Am J Hum Genet 80:605–615
Conflict of interest
The authors declare no conflict of interest related to this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ntzani, E.E., Liberopoulos, G., Manolio, T.A. et al. Consistency of genome-wide associations across major ancestral groups. Hum Genet 131, 1057–1071 (2012). https://doi.org/10.1007/s00439-011-1124-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1124-4