Abstract
An efficient method to uniquely identify every individual would have value in quality control and sample tracking of large collections of cell lines or DNA as is now often the case with whole genome association studies. Such a method would also be useful in forensics. SNPs represent the best markers for such purposes. We have developed a globally applicable resource of 92 SNPs for individual identification (IISNPs) with extremely low probabilities of any two unrelated individuals from anywhere in the world having identical genotypes. The SNPs were identified by screening over 500 likely/candidate SNPs on samples of 44 populations representing the major regions of the world. All 92 IISNPs have an average heterozygosity >0.4 and the F st values are all <0.06 on our 44 populations making these a universally applicable panel irrespective of ethnicity or ancestry. No significant linkage disequilibrium (LD) occurs for all unique pairings of 86 of the 92 IISNPs (median LD = 0.011) in all of the 44 populations. The remaining 6 IISNPs show strong LD in most of the 44 populations for a small subset (7) of the unique pairings in which they occur due to close linkage. 45 of the 86 SNPs are spread across the 22 human autosomes and show very loose or no genetic linkage with each other. These 45 IISNPs constitute an excellent panel for individual identification including paternity testing with associated probabilities of individual genotypes less than 10−15, smaller than achieved with the current panels of forensic markers. This panel also improves on an interim panel of 40 IISNPs previously identified using 40 population samples. The unlinked status of the subset of 45 SNPs we have identified also makes them useful for situations involving close biological relationships. Comparisons with random sets of SNPs illustrate the greater discriminating power, efficiency, and more universal applicability of this IISNP panel to populations around the world. The full set of 86 IISNPs that do not show LD can be used to provide even smaller genotype match probabilities in the range of 10−31–10−35 based on the 44 population samples studied.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In previous papers (Kidd et al. 2006; Pakstis et al. 2007), we described the rationale and our strategy for developing a panel of SNPs for individual identification (IISNPs) and presented some potentially useful IISNPs. Such a panel would have use in sample tracking in large collections of human DNA samples and in forensics and paternity testing. Others have also addressed the value of such panels in forensics (Inagaki et al. 2004; Lee et al. 2005; Sanchez et al. 2006; Butler et al. 2008; Pakstis et al. 2008). One panel of 52 SNPs has been accepted for forensic use in several European countries (Sanchez et al. 2006; Phillips et al. 2009). An IISNP panel would provide a complementary tool for forensic applications in situations, such as highly degraded DNA (e.g., Fang et al. 2009), in which the standard STR markers of the widely used COmbined DNA Index System (CODIS) panel do not perform well. SNPs also offer a potentially cheaper, faster, and more automatable alternative to STRs in many applications. While any sufficiently large set of SNPs will guarantee uniqueness of every individual, there are clear advantages to a set with extensive population genetic support and standardization, if possible, to allow comparability between groups and studies. In the interest of efficiency, we have defined criteria for an IISNP panel: the SNPs should have very little variation in frequency across human populations and be highly informative around the world as measured by F st and expected heterozygosity, respectively (Kidd et al. 2006). We have arbitrarily chosen a global F st < 0.06 and global average heterozygosity >0.4. A sufficient number of SNPs is needed so that the average match probabilities (the probabilities of two unrelated individuals having the same multi-locus genotype) of the final panel should at least be comparable to the standard CODIS STR markers (Budowle et al. 1998). An interim report (Pakstis et al. 2007) of our progress in developing an IISNP panel documented 40 SNPs meeting these criteria based on 40 population samples representing the major continental regions of the world. Short reports (Butler et al. 2008; Pakstis et al. 2008) described aspects of the IISNP search as well as discussions of the potential role of IISNPs in forensic applications. We have since revised our criteria to require that a final core panel of markers would be unlinked in order to make them more generally useful, especially in identification scenarios involving close biological relatives and in paternity testing.
In our original study, we described a strategy based on having data available a priori on only a very few populations. Recently high throughput SNP dataset resources involving many different populations have become available for identification of appropriate candidate SNPs: 14 populations studied by Shriver et al. (2005) and studies of the HGDP-CEPH panel of 52 populations (Li et al. 2008; Conrad et al. 2006; Pemberton et al. 2008). The availability of these resources has allowed a marked improvement in the efficiency of our search for additional IISNPs. We scanned those datasets targeting regions of the human autosomal genome in which we had not previously found useful markers in order to find additional unlinked SNPs meeting our criteria. Therefore, our search uncovered a large number of additional SNPs with the desired population genetic properties and better molecular distributions. We also were able to expand our set of test populations by adding four groups from geographic regions poorly represented in the initial 40 populations.
Our final SNP panel for individual identification consists of 86 IISNPs that meet our criteria based on samples of 44 populations representing the major human populations around the world and includes a subset of 45 unlinked SNPs that provide match probabilities in these 44 populations that are at least comparable to and sometimes better than the standard CODIS STR markers.
Methods
Our previous publications (Kidd et al. 2006; Pakstis et al. 2007) described the strategy and goal for developing a panel of IISNPs. Briefly, we have identified in publically available population data SNPs that were likely to meet the criteria and then screened them on our much larger set of 44 populations. The core criteria for accepting an IISNP remain unchanged in that each SNP must have an average heterozygosity ≥0.4 for all the populations studied and the F st value across those populations must be <0.06. All candidate IISNPs, including the 40 previously published (Pakstis et al. 2007) were typed and evaluated on all 44 population samples. The recent selection of candidates preferentially targeted chromosomal regions that had not yet produced IISNPs in order to maximize the number of SNPs that would be essentially unlinked.
Table s1 of the supplemental material lists all 44 population samples studied along with their unique population and sample identifiers (UIDs) in the ALlele FREquency Database (ALFRED, http://alfred.med.yale.edu), where details on each population and sample are described. The four new population samples added to the set of 40 populations already described (Pakstis et al. 2007) are Sandawe from Tanzania (40), Hungarians (92), Keralites from Southern India (30), and Laotians (119).
All SNPs screened were typed by TaqMan® using assays obtained from Applied Biosystems. All reactions were done in 384-well plates in 3 μl reactions and then read on an AB7900HT with interpretations by SDS software (version 2.3) augmented by visual inspection of the clustering to insure conservative interpretations.
The allele frequencies for each SNP were estimated by gene counting within each group studied assuming each marker is a two-allele co-dominant genetic system. The polymorphisms were tested for agreement with Hardy–Weinberg ratios in each population sample studied by comparing the expected and observed number of individuals occurring for each possible genotype in a simple Chi-square test. In the few cases, in which a number for a particular genotype was small the statistical significance was evaluated by a Monte Carlo based permutation procedure employing 1,000 iterations (Cubells et al. 1997).
The chromosome nucleotide position shown in Table 1 for each SNP follows Genome Build 36.2. The genetic map position in centi-Morgans (cM) was determined for each SNP by computing a simple average of the interpolated DeCode, Genethon, and Marshfield genetic map values obtained for each polymorphism by entering the nucleotide position into the NCBI Map Viewer and recording the values reported for each reference map. The starting or zero map position is assumed to be near the pter end of each chromosome. Each of these extensive genetic maps does not necessarily have the same starting point on each chromosome and the density of markers will vary in different chromosome regions. These nucleotide positions and approximate genetic map distances were employed in the process of selecting the subset of 45 unlinked IISNPs. A reviewer of this manuscript brought to our attention the existence of another valuable human genetic map based on over 28,000 markers (SNPs and STRPs) available online—the Rutgers Map—(Matise et al. 2007). We compared the interpolated centi-Morgan map distances provided by the Rutgers Map with the average genetic map values for each SNP in Table 1 and found them to be very similar (mean 3 cM difference). Thus, they reinforce the decisions made earlier based on the three maps available via the NCBI map viewer.
In order to evaluate the statistical independence of the SNPs, linkage disequilibrium values, r 2 (Devlin and Risch 1995) were computed for all unique pairings of the 92 SNPs in each population sample. The LD values were screened in a variety of ways to determine whether there was any evidence for meaningful associations among the markers.
Match probabilities and most common multi-locus genotype frequencies were calculated as previously described (Kidd et al. 2006). Hardy–Weinberg ratios and the statistical independence of the loci were assumed.
Results
We screened over 500 SNPs that appeared to be likely candidates meeting our criteria based on information such as estimated allele frequencies from publically available data. Table 1 presents the final list of 92 IISNPs that our study identified as individually meeting our F st and heterozygosity criteria. The SNPs are ranked in ascending order according to the F st value for the 44 population samples studied. In the case of SNPs with identical F st values, the SNP with the higher average heterozygosity was assigned the lower/better rank. The 45 unlinked SNPs are also indicated. A more detailed, annotated version of Table 1 can be found as a pdf file at (http://info.med.yale.edu/genetics/kkidd/92snpJan2009.pdf). We have deposited in ALFRED the allele frequencies and samples sizes for all population samples and all SNPs screened in this project including those that were not included among the final 92 IISNPs.
No meaningful deviations from Hardy–Weinberg ratios occur for any of the 92 IISNPs in the 44 population samples. For the 92 × 44 = 4,048 tests the proportion of probabilities obtained falling below the 5, 1, and 0.1% significance level thresholds (1.88, 0.27, and 0.05% respectively) were generally somewhat smaller than the values expected by chance due in part to the extensive selection procedure that included discarding candidate SNPs with strong Hardy–Weinberg deviations. Moreover, the population samples had previously been tested for large numbers of SNPs as part of other studies and were expected to show no systematic deviations from Hardy–Weinberg ratios.
Pairwise LD calculations for all 92 IISNPs show that removal of 6 IISNPs with very close linkage (those with ranks 52, 57, 65, 66, 68, and 89 in Table 1) leaves 86 IISNPs with no significant pairwise LD across the populations. Among the 160,820 tests of LD for all possible pairings of 86 SNPs, there remain 7 nominally significant LD values ranging from 0.40 to 0.69 that display no obvious pattern and are likely due to chance: 6 of these 7 outliers involve pairings of SNPs on different chromosomes, each involving a different pair of SNPs in a different population. The seventh pair involves SNPs more than 161 MB apart on the same chromosome. Additional details are presented in the Supplemental Material.
Among the 86 IISNPs, we identified a set of 34 markers that have essentially zero linkage because they are either on separate chromosomes or are separated by distances greater than 95 cM (roughly the centiMorgan distance that with a Kosambi correction would give 50% recombinant gametes). An additional 11 IISNPs are separated from any of the other IISNPs that are syntenic by map distances of 41–94 cM indicating loose to almost no linkage. We consider this subset of 45 IISNPs to constitute an unlinked panel for practical purposes. There are multiple additional SNPs among the remaining 47 IISNPs in Table 1 that could be substituted for some of the 45 without greatly altering the essential absence of linkage.
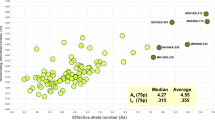
This recommended subset of 45 unlinked IISNPs has exceptional information content (median heterozygosity = 0.478 and 93.2% of the 1,980 individual heterozygosity values ≥0.4). When pairwise LD does not exist, as among these 45 unlinked IISNPs as well as among the remainder of the 86 SNPs, the SNPs are statistically independent at the population level and the “product rule” can be used to calculate match probabilities. Figure 1 displays match probabilities and the most common genotype frequencies for each population for our recommended set of 45 unlinked IISNPs using the actual allele frequency estimates for each population. Most of the populations have match probabilities <10−17 and many are <10−18; the smaller, more isolated populations still have match probabilities <10−15. Thus, this set of 45 unlinked SNPs is an excellent panel for individual identification with match probabilities comparable to the CODIS STR panel. Another desirable characteristic is that the probabilities are essentially independent of ethnicity since allele frequency differences between populations are so small. Consequently, it is conservative to say with considerable scientific justification that a maximum match probability of <10−15 can be used for the probability that any two individuals from anywhere in the world will have identical genotypes. The unlinked status of these 45 SNPs also makes them useful for situations involving close biological relationships. In paternity testing, the much lower probability of mutations occurring at SNPs relative to STRPs makes SNPs useful in general and these IISNPs are especially informative. If biological relationships are not involved, more of the 86 IISNPs can be added to the set to make the match probabilities even smaller. Computing match probabilities based on all 86 IISNPs that show no pairwise LD gives values in the range of 10−31–10−35 for the 44 populations.
Match probabilities and the most common genotype frequencies for each of the 44 population samples based on the 45 unlinked SNPs. Populations are ordered from African on the left, through the Middle Eastern, European, Central Asian, Siberian, Pacific Islanders, and East Asian, to Native American on the right
Discussion
We have identified an improved panel of 86 SNPs that individually have high heterozygosity combined with very low F st for the worldwide sampling of populations studied. This set of IISNPs has no significant linkage disequilibrium between any pair in any of the populations so that each SNP could be considered to be statistically independent at the population level. Even though a few large LD values occur, as noted in Results, they are not meaningful. Moreover, the outlier LD values above any arbitrary threshold, such as 0.2 and 0.3, typically involve populations with relatively small sample sizes. The bias toward larger LD values that occurs when sample sizes are small is a known phenomenon and was discussed in our previous report (Pakstis et al. 2007). The correlation coefficient between sample size (2n) and LD values for the whole dataset equals −0.23. A subset of 45 SNPs also shows no close linkage between any pair of SNPs so that they are also statistically independent in situations involving biological relationships. The enlarged set of 44 representative world populations (supplemental Table s1) has increased the stringency of the inclusion criteria over the preliminary panel reported previously (Pakstis et al. 2007). Because many of those previous 40 IISNPs showed significant linkage, only 23 of them are among the present set of 45 unlinked IISNPs.
Additional optimization of population characteristics and spacing of markers is possible. However, we have settled on the current panel of 45 “unlinked” IISNPs. Even though 11 of the SNPs show very loose linkage to any of the others, the statistical consequences are minimal. In individual family situations, the statistics assuming no linkage will be minimally different from those using estimates from the linkage map. Moreover, a linkage parameter becomes relevant only when dealing with double heterozygotes; they occur at only a maximum of 25% of the time for any specific pair of loci. We believe that additional effort at optimization is not warranted by the slight improvement that would presumably be possible.
The average probabilities of two individuals from anywhere in the world having identical genotypes for the 45 IISNPs in Fig. 1 are all below 10−15 compared to 10−13 for the 40 best SNPs in Pakstis et al. (2007) and 38 of the 44 populations have such match probabilities less than 10−17 in a range typical of the best that can be achieved with CODIS markers in populations with higher heterozygosity. That this is an efficient set of IISNPs is illustrated in Fig. 2 by comparisons with two “random” sets of SNPs. The two sets of 45 non-overlapping random SNPs are distributed across most of the autosomes and derive from a collection of more than 4,000 SNPs unselected for forensic purposes and typed on the 44 population samples. The ~4,000 SNPs were mostly selected for variability in most of the world’s major geographical regions but they were selected neither for high heterozygosity nor for low F st.
Comparison of match probabilities for the 45 unlinked IISNPs with two different, non-overlapping random samplings of 45 SNPs in the 44 population samples. The match probabilities for the 45 IISNPs are substantially better (smaller), on average many orders of magnitude better, than those provided by the random SNP sets. There is also greatly reduced variability in average match probabilities across the 44 populations for the 45 unlinked IISNPs compared to either of the random SNP sets, especially outside of Europe
These comparisons empirically demonstrate the value of the screening process we have followed in developing the IISNP panel. We recognize that any sufficiently large set of random SNPs could achieve the same low match probabilities, but argue that a single efficient set has value, especially for tracking samples shared between laboratories. Because the raw data have been public at our website since January 2009, we know that Applied Biosystems has already begun developing multiplex reactions now in beta test. Other companies may also be pursuing such kits. This could allow samples in many different labs to be uniquely identified by a common “bar code” system.
While the screening process identifying the 92 IISNPs has resulted in a set of SNPs each of which has very similar frequencies across the 44 population samples studied, small differences in gene frequencies arising from some combination of real and sampling noise variation are still observed across the ethnic groups studied. To assess how much predictability for ethnicity might exist in the set of IISNPs, we have carried out a series of analyses with the STRUCTURE program version 2.3.1 (Pritchard et al. 2000) using the standard admixture model to compare the 92 IISNPs with a set of 200 random SNPs on the 44 population samples. No noticeable predictability of ethnicity (population structure) can be achieved with the 92 IISNPs for a series of analyses specifying 2 through 10 clusters (K values). In strong contrast, 200 random SNPs can be useful in predicting the continental membership of an individual (see Supplemental Material).
Principal components analysis (PCA) provides another way of visualizing the structure present. Figure 3 presents scatter plots of PCA analyses on the 44 population samples for two sets of SNPs—the 92 IISNPs (Fig. 3a) and 200 random SNPs (Fig. 3b). The input files for the PCA analyses consisted of the tau genetic distance matrices computed from the SNP allele frequencies. The results of the first two principal components of their respective analyses are plotted in each figure. Figure 3a based on the IISNPs accounts for 41% of the variation and most of the populations can be seen to cluster closely together in the center of the figure with the primary differentiation visible arising from the samples of relatively small, inbred populations that define the distal points of the plotted axes. In strong contrast, Figure 3b based on the random SNPs accounts for about 72% of the variation and the population samples can be seen to group into very clear geographical clusters corresponding to the major continents. While Fig. 3a does still display some residual indications of geographical clustering when examined closely, they are very weak compared to the strong, distinct clustering based on the random SNPs.
The 92 IISNPs in Table 1 also meet another important criterion beyond the purely population genetic ones. No medical or sensitive personal information is conveyed by the individual or combined data. To our knowledge, these SNPs are not in any “gene” or other type of functional element other than protein coding sequences. That does not eliminate the possibility that a functional difference will be identified for alleles at one or more of the IISNPs. However, since these SNPs approach the ideal of 50% heterozygosity, an average of about 37.5% of the global human population will share any randomly chosen genotype at any one of the loci. That minimizes the level of concern should some functional effect of one of these SNPs be determined in the future since all genotypes must be considered normal.
Our final set of 86 IISNPs that have no significant LD has excellent characteristics that qualify it for being accepted as a universal panel for individual identification. The 45 unlinked IISNPs already yield match probabilities that come close to the theoretical average match probability of just under 10−19 for 45 “perfect” IISNPs, i.e., all with heterozygosity equal to 0.5. While our use of F st < 0.06 is arbitrary, it has proven to be very good at identifying markers with very similar allele frequencies in most populations. As more populations are typed, especially smaller and/or more isolated populations, some of these 45 SNPs may have less uniformly high heterozygosities. Certainly, their rank order is expected to change when any additional populations are considered. However, it is extremely unlikely that match probabilities for the 45 unlinked SNPs will exceed 10−12, still a very meaningfully low value. In addition, with 86 SNPs independent at the population level, some of which could be substituted for some of the 45 unlinked SNPs should technical (e.g., multiplexing) problems arise; we think that pursuit of additional IISNPs will not be necessary.
For actual applications that employ either the 45 unlinked IISNPs or the full 86 IISNP panel, we have assumed that users will include various additional markers for such purposes as quality control (such as duplicating some SNPs) and identification of gender (for example, the amelogenin gene, AMELX, marker already in standard use in forensic studies). There should be ample room in standard 96-well formats to accommodate such additional markers.
References
Budowle B, Moretti TR, Niezgoda SJ, Brown BL (1998) CODIS and PCR-based short tandem repeat loci: law enforcement tools. In: Second European symposium on human identification, Promega Corporation, Madison
Butler JM, Budowle B, Gill P, Kidd KK, Phillips C, Schneider PM, Vallone PM, Morling N (2008) Report on ISFG SNP Panel Discussion. In: Progress in forensic genetics: genetics supplement series, vol 1, pp 471–472
Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK (2006) A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat Genetics 38:1251–1260
Cubells JF, Kobayashi K, Nagatsu T, Kidd KK, Kidd JR, Calafell F, Kranzler H, Ichinose H, Gelernter J (1997) Population genetics of a functional variant of the dopamine beta-hydroxylase gene (DBH). Am J Med Genetics Neuropsych Genet 74:374–379
Devlin B, Risch N (1995) A comparison of linkage disequilibrium measures for Wne-scale mapping. Genomics 29:311–322
Fang R., Pakstis AJ, Hyland F, Wang D, Shewale J, Kidd JR, Kidd KK, Furtado MR (2009) Multiplexed SNP detection panels for human identification. Forensic Sci Int Gene Suppl (in press). doi:10.1016/j.fsigss.2009.08.161
Inagaki S, Yamamoto Y, Doi Y, Takata T, Ishikawa T, Imabayashi K, Yoshitome K, Miyaishi S, Ishizu H (2004) A new 39-plex analysis method for SNPs including 15 blood group loci. Forensic Sci Int 144:45–57
Kidd KK, Pakstis AJ, Speed W, Grigorenko E, Kajuna SLB, Karoma N, Kungulilo S, Kim J-J, Lu A, Odunsi R-B, Okonofua F, Parnas J, Schulz L, Zhukova O, Kidd JR (2006) Developing a SNP panel for forensic identification of individuals. Forensic Sci Int 164:20–32
Lee HY, Park MJ, Yoo J-E, Chung U, Han G-R, Shin K-J (2005) Selection of twenty-four highly informative SNP markers for human identification and paternity analysis in Koreans. Forensic Sci Int 148:107–112
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM (2008) Worldwide human relationships inferred from genome-wide patterns of variation. Science 319:1100–1104
Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, Hyland FCL, Kennedy GC, Kong X, Murray SS, Ziegle JS, Stewart WCL, Buyske S (2007) A second-generation combined linkage-physical map of the human genome. Genome Res 17:1783–1786
Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2007) Candidate SNPs for a universal individual identification panel. Hum Genet 121:305–317
Pakstis AJ, Speed WC, Kidd JR, Kidd KK (2008) SNPs for individual identification. In: Progress in forensic genetics: genetics supplement series, vol 1, pp 479–481
Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA (2008) Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Ann Hum Genet 72:535–546
Phillips C, Prieto L, Fondevila M, Salas A, Gomez-Tato A, Alvarez-Deos J, Alonso A, Bianco-Verea A, Brion M, Montesino M, Carracedo A, Lareu MV (2009) Ancestry analysis in the 11-M Madrid bomb attack investigation. PLOS ONE 4:e6583
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Sanchez JJ, Phillips C, Borsting C, Balogh K, Bogus M, Fondevila M, Harrison CD, Musgrave-Brown E, Salas A, Syndercombe-Court D, Schneider PM, Carracedo A, Morling N (2006) A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 27:1713–1724
Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, Friedlaender J, Jorde LB, Watkins WS, Bamshad MJ, Guiterrez G, Loi H, Matsuzaki H, Kittles RA, Argyropoulos G, Fernandez JR, Akey JM, Jones KW (2005) Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics 2:81–89
Acknowledgments
This work was funded primarily by NIJ Grants 2004-DN-BX-K025 and 2007-DN-BX-K197 to KKK awarded by the National Institute of Justice, Office of Justice Programs, US Department of Justice. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the US Department of Justice. We thank Applied Biosystems for making their allele frequency database available to us and for supplying some of the TaqMan reagents that were employed in these studies. We also thank Eva Straka for excellent technical help. We also want to acknowledge and thank the following people who helped assemble the population samples from the diverse populations over a period of many years: C. Barta, F. L. Black, B. Bonne-Tamir, L. L. Cavalli-Sforza, K. Dumars, J. Friedlaender, L. Giuffra, E. L. Grigorenko, S. L. B. Kajuna, N. J. Karoma, K. Kendler, J.-J. Kim, W. Knowler, S. Kungulilo, H. Li, R.-B. Lu, A. Odunsi, F. Okonofua, F. Oronsaye, J. Parnas, L. Peltonen, H. Rajeevan, L. O. Schulz, D. Upson, K. Weiss, and O. V. Zhukova. In addition, some of the cell lines were obtained from the National Laboratory for the Genetics of Israeli Populations at Tel Aviv University, Israel, and the African American samples were obtained from the Coriell Institute for Medical Research, Camden, NJ. Special thanks are due to the many hundreds of individuals who volunteered to give blood samples for studies of gene frequency variation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pakstis, A.J., Speed, W.C., Fang, R. et al. SNPs for a universal individual identification panel. Hum Genet 127, 315–324 (2010). https://doi.org/10.1007/s00439-009-0771-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-009-0771-1