Abstract
Fruit trees, such as apple (Malus × domestica Borkh.), are woody perennial plants with a long juvenile phase. The biological analysis for the regulation of flowering time provides insights into the reduction of juvenile phase and the acceleration of breeding in fruit trees. In Arabidopsis, LIKE HETEROCHROMATIN PROTEIN1 (LHP1) is involved in epigenetic silencing of the target genes such as flowering genes. We isolated and characterized twin apple LHP1 homolog genes, MdLHP1a and MdLHP1b. These genes may have been generated as a result of ancient genome duplication. Although the putative MdLHP1 proteins showed lower similarity to any other known plant LHP1 homologs, a chromo domain, a chromo shadow domain, and the nuclear localization signal motifs were highly conserved among them. RT-PCR analysis showed that MdLHP1a and MdLHP1b were expressed constantly in developing shoot apices of apple trees throughout the growing season. Constitutive expression of MdLHP1a or MdLHP1b could compensate for the pleiotropic phenotype of lhp1/tfl2 mutant, suggesting that apple LHP1 homolog genes are involved in the regulation of flowering time and whole-plant growth. Based on these results, LHP1 homolog genes might have rapidly evolved among plant species, but the protein functions were conserved, at least between Arabidopsis and apple.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transition from juvenile to adult phase in plant species requires environmental cues, such as temperature, light intensity, and day length, and internal cues, such as phytohormones, age, and body size and thus the perennial fruit trees, such as apple (Malus × domestica Borkh.), have a long juvenile phase lasting about 4-8 years (Hackett 1985; Dennis Jr 2003). The long juvenile phase is the primary factor that limits the efficient breeding and the genetic and molecular analyses of fruit trees. Therefore, the studies of the molecular basis of flower differentiation would provide insights into the solution of such issues in the area of horticulture. Because apple is one of the most economically important fruit tree crops in the world, many researchers have studied this one genetically and physiologically from the view point of both industry and scientific interest. A decade ago, molecular studies on the morphogenesis in apple also started and several genes related to plant development have been isolated and characterized. Recent studies on the genetic control of flowering time in apple suggest that MdTFL1, a TERMINAL FLOWER1(TFL1)-like gene of apple, is a key regulatory gene involved in the maintenance of the juvenile and vegetative phase in apple (Kotoda et al. 2006). However, little is known about other genes regulating the flowering time of apple, and much less about the genetic framework of the life cycle of apple.
On the other hand, studies on Arabidopsis thaliana have led to the identification of many genes controlling the flowering time. Previous studies revealed that some mutants lacking the genes encoding Polycomb group (PcG) proteins or chromatin-remodeling factors show early flowering. CURLY LEAF (CLF) was first identified as a PcG gene in plant, which represses AGAMOUS (AG) expression (Goodrich et al. 1997). The EMBRYONIC FLOWER2 (EMF2) encoding a PcG protein with a zinc finger motif is required for maintaining the vegetative development and regulating the development of the inflorescence and floral meristem (Yoshida et al. 2001). These PcG family proteins form large multimeric complexes and regulate developmental processes by preventing inappropriate expression of target genes (Chanvivattana et al. 2004). For example, CLF/SWINGER (SWN), EMF2, FERTILISATION-INDEPENDENT ENDOSPERM (FIE), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) constitute a PcG complex, which predominantly maintains AG expression in a transcriptionally repressed state (Chanvivattana et al. 2004).
Arabidopsis LIKE HETEROCHROMATIN PROTEIN1 (LHP1) [TERMINAL FLOWER2 (TFL2)] is a single-copy gene encoding a homolog protein of HETEROCHROMATIN PROTEIN 1 (HP1), which is composed of two characteristic domains: an N-terminal chromatin organization modifier (chromo) domain (CD) and a C-terminal chromo shadow domain (CSD) (Gaudin et al. 2001; Kotake et al. 2003). The HP1-family proteins are known to regulate chromatin function from mammals to yeast, and may act similarly as PcG proteins by silencing genes (Maison and Almouzni 2004). Animals have three isoform proteins, HP1α, HP1β, and HP1γ, whereas plants harbor a single-copy gene encoding a homologous protein more related to HP1γ according to its euchromatin localization (Minc et al. 2000; Zemach et al. 2006). The loss-of-function lhp1/tfl2 allelic mutants showed the pleiotropic phenotypes, such as early flowering, terminal flower, small plant size, and curled leaf (Larsson et al. 1998; Gaudin et al. 2001; Kotake et al. 2003). In addition, the double mutants of tfl1 and tfl2 showed a more severe phenotype than that of a single mutant (Larsson et al. 1998). DNA microarray analysis in the tfl2 mutant showed that the expression levels of euchromatin genes, such as FLOWERING LOCUS T (FT), which is a flowering integrator gene, and APETALA3, PISTILLATA, AG, and SEPALLATA3, which are floral homeotic genes, are up-regulated (Kotake et al. 2003; Takada and Goto 2003; Nakahigashi et al. 2005; Germann et al. 2006). Thus, the pleiotropic phenotypes of the lhp1/tfl2 mutants are likely to be reflected by the fact that LHP1 represses the expression of multiple genes.
In this study, we report the isolation and characterization of two LHP1 homolog genes in apple, MdLHP1a and MdLHP1b. These genes showed lower identity to other known LHP1 homologs in plant species, but constitutive expression of MdLHP1a or MdLHP1b rescued the Arabidopsis lhp1/tfl2 mutant. Consequently, LHP1 homolog genes in the apple of woody perennial may also be involved in the regulation of the flowering pathway, and conserved functionally in plant species. In addition, these apple LHP1 homolog genes may have been duplicated because the Maloideae subfamily possesses the complex genome derived from the origin of polyploidy.
Materials and methods
Plant materials
Different organ tissues and shoot apices of apple (Malus × domestica Borkh.) cultivars (cv.) ‘Fuji’ and ‘Jonathan’ were collected from the experimental field at the National Institute of Fruit Tree Science in Morioka, Japan. These cultivars were used for the seasonal and the tissue specific expression analyses because they are popular cultivars and the genetic background is not so different between them. The seedlings from ‘Fuji’ apple were used for the expression analysis of MdLHP1a and MdLHP1b in the juvenile phase. The A. thaliana tfl2-2 loss-of-function mutant was obtained from the Arabidopsis Biological Resource Center at the Ohio State University and employed for the mutant rescue experiment.
cDNA library construction and cloning of MdLHP1a and MdLHP1b
Total RNA was isolated from flower buds of ‘Fuji’ apple collected on April 26, 2004, in Morioka, Japan, with the CTAB method as described by Kotoda et al. (2000). Poly (A)+ RNA was isolated using a PolyA tract mRNA purification kit (Promega, Madison, WI, USA). The cDNA library was constructed using a ZAP-cDNA Synthesis Kit (Stratagene, La Jolla, CA, USA) and Packagene® Lambda DNA Packaging System (Promega). The average size of the inserted cDNAs was about 1.3 kbp. PCR amplification of Arabidopsis LHP1 DNA fragments was carried out with a pair of primers (LHP1F: 5′-TAG GAA ACG GAA GCG CAA ATA TGC AG-3′; LHP1R: 5′-CCA ACA CTT CCT GGA CAT TGT CTG AT-3′). The PCR-amplified fragment (gLHP1, 696 bp) was cloned into the pT7Blue vector (Novagen, Darmstadt, Germany). The gLHP1 labeled with [α-32P]dCTP (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) was used as a probe to screen 1.0 × 105 pfu of the apple cDNA library. A positive plaque was excised to the pBluescript SK(−) phagemid clone (MdLHP1a1), and this clone was then sequenced. Then, 5.0 × 105 pfu of the cDNA library was screened with a MdLHP1a1 probe labeled with digoxigenin (DIG, Roche Diagnostics, Mannheim, Germany), which was amplified by PCR with a pair of primers designed for the 5′ region (MdLHP1F-NheI: 5′-GTA GCT AGC ACA AAT GAG AAC CAA GG-3′ with NheI site in bold and ATG in italic; MdLHP1-600R: 5′-GAC GAT GCC AAT TTC CAC AT-3′). The hybridization was performed in DIG Easy Hyb (Roche Diagnostics) at 42°C for 16 h followed by two rinses in 2× SSC containing 1% (w/v) SDS at room temperature for 5 min and two washes in 0.5× SSC containing 1% (w/v) SDS at 68°C for 20 min. Chemiluminescent signals were visualized using the LAS1000 image analyzer (Fuji Photo Film, Aichi, Japan). PCR amplification of MdLHP1a genomic DNA was carried out in a mixture of a pair of primers [MdLHP1F-NheI and MdLHP1a1R-XhoI: 5′-TGC CTC GAG ACT AAA ATG TAG GGT TGT AT-3′ with XhoI site in bold and CTA (stop codon) in italic], EX-Taq DNA polymerase (Takara Biomedical, Tokyo, Japan), and 250–300 ng of genomic DNA as a template. PCR was programmed for preheating at 96°C for 3 min followed by 30 cycles of 96°C for 20 s, 58°C for 30 s, and 72°C for 5 min. A 4.6-kb PCR-amplified product was cloned into the pT7Blue vector (Novagen).
Sequence analyses
The nucleotide sequence was determined using a DTCS Quick Start Kit for Dye Terminator Cycle Sequencing (Beckman Coulter, Fullerton, CA, USA) and an automated DNA sequencer CEQ 8000 (Beckman Coulter). Nucleotide and amino acid sequences were analyzed using CLUSTAL X multiple sequence alignment program Version 1.83 (Jeanmougin et al. 1998) and GeneDoc (Nicholas et al. 1997). The phylogenetic tree was displayed using the Njplot program (Perrière and Gouy 1996). A homology search was performed with BLAST2 of the GenomeNet (http://www.genome.ad.jp/).
Southern blot analysis
Southern blot analysis was carried out according to the method described by Sambrook et al. (1989). Fifteen micrograms of genomic DNA, isolated from the leaves of apple, was digested with EcoRI, PstI, BamHI, or EcoRV and then electrophoresed on a 0.8% agarose gel. The DNA bands were blotted onto a nylon membrane Hybond-N+ (GE Healthcare Bio-Sciences Corp.) and hybridized with the full-length cDNA of MdLHP1a1 labeled with DIG by PCR as a probe. The hybridization signals were detected in the same manner as described in the section of cloning.
Expression analyses by RT-PCR and Northern hybridization
Total RNAs were extracted from flower buds (late April), fruit receptacles, and apical buds of fruit-bearing shoots (FBS) and succulent shoots (SS) (from June to October in 2004) of adult ‘Jonathan’ apple trees and stems, apical buds of vegetative shoots (VS), and leaves of 1-month-old seedlings and cotyledons and roots of 6-day-old seedlings. For RT-PCR analysis, first-strand cDNAs were synthesized from 2 μg of total RNAs in 30 μl of reaction mixture using the ReverTra Ace kit (Toyobo, Tokyo, Japan). The subsequent PCR reactions were performed with 0.5 μl of the first-strand cDNA reaction mixture as templates. A fragment of MdLHP1a1/-a2 or MdLHP1b1/-b2 was amplified with the following primers: sense primer (5′-CAG CAG CGT TCT ACT GAT CCT ATT TAC AAT-3′) and antisense primer (5′-GAT CCA GAT ACA AAT CTC ATA ATA TTT CCT AAT ACA G-3′) for MdLHP1a1/-a2 and sense primer (5′-CAG AGC AAT CGC AGC ACA CTG GA-3′) and antisense primer (5′-CTC ATC CAA ACT CCG ACT AGT CCC-3′) for MdLHP1b1/-b2. As an internal control, a fragment of apple Histone H3 was amplified with the following primers: sense primer (5′-TGA AGA AGC CCC ACA GAT A-3′) and antisense primer (5′-ACA CAA GAA ACT ATA AAC C-3′) as described by Kotoda et al. (2006). Thermal cycle programs were as follows: 96°C for 3 min followed by 40 cycles of 96°C for 20 s, 65°C for 30 s, and 72°C for 3 min for MdLHP1a1/-a2 and MdLHP1b1/-b2 and 96°C for 3 min followed by 32 cycles of 96°C for 20 s, 56°C for 30 s, and 72°C for 30 s for Histone H3. For Northern analysis, total RNA from wild-type and transgenic Arabidopsis plants was electrophoresed on a denaturing 1.2% agarose gel with formaldehyde and then blotted onto a Hybond N+ membrane (GE Healthcare Bio-Sciences Corp.). The blotted membrane was hybridized with a DIG-labeled MdLHP1a1 probe. The hybridization was performed in DIG Easy Hyb at 50°C for 16 h. The wash and detection were performed in the same manner as that used for DNA.
Construction of transformation vector
To construct a vector for the constitutive expression of MdLHP1a, the coding region of MdLHP1a1 was amplified by PCR with a pair of primers MdLHP1F-NheI and MdLHP1a1R-XhoI. The PCR products were subsequently digested with NheI and XhoI and then cloned into the XbaI–SalI sites of the plasmid vector pBI9526Ω (a derivative of pBI221) bearing the cauliflower mosaic virus (CaMV) 35S promoter fused with the Ω sequence (Gallie and Walbot 1992) and the nopaline synthase terminator (Tnos) (designated as 35SΩ:MdLHP1a1:Tnos/pBI9526Ω). After 35SΩ:MdLHP1a1:Tnos/pBI9526Ω was digested with EcoRI, the DNA fragment of 35SΩ:MdLHP1a1:Tnos was cloned into the respective sites of the pSMAK312Blue binary vector. The binary vector obtained was named 35SΩ:MdLHP1a1. To construct a vector for constitutive expression of MdLHP1b, the coding region of MdLHP1b1 was amplified with a pair of primers MdLHP1F-NheI and MdLHP1b1R-KpnI [5′-ATC GGT ACC TTA AAA TGT AGG ATT GTA TCG-3′ with KpnI site in bold and TTA (stop codon) in italic]. It was then cloned into the XbaI–KpnI sites of modified pSMAK193E (35SΩ/pSMAK193E) to be placed between the promoter 35SΩ and the terminator of the Arabidopsis rubisco gene (TrbcS). The resultant binary vector was named 35SΩ:MdLHP1b1. The pSMAK binary vector series, pSMAK312Blue and pSMAK193E, were made from the pSMAK251 vector. The characterization of pSMAK251, which contains genes encoding β-glucuronidase (GUS) and neomycin phosphotransferase II (NPTII) in its T-DNA region, will be published elsewhere (H. Ichikawa, in preparation).
Arabidopsis transformation
Wild-type (ecotype Columbia) and tfl2-2 Arabidopsis plants were transformed with 35SΩ:MdLHP1a1 and 35SΩ:MdLHP1b1 using an Agrobacterium tumefaciens strain, EHA101 (Hood et al. 1986), by the infiltration method as described by Bechtold et al. (1993). Bleach-sterilized seeds were place on MS plates (Murashige and Skoog 1962) containing carbenicillin (250 mg/l) and kanamycin (35 mg/l). Arabidopsis seeds of the T3 plant (primary transformants were defined as T1 plants) were sown on agar plates followed by stratification at 4°C for 3 days and grown in a growth chamber controlled at 22°C under long-day (LD) conditions (16 h light/8 h darkness) or short-day (SD) conditions (8 h light/16 h darkness).
Results
Isolation of four cDNAs of LHP1 homolog genes from apple
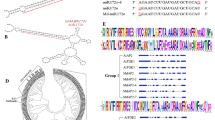
To isolate the apple LHP1 homolog genes, the PCR-amplified DNA fragment (696 bp) of Arabidopsis LHP1 was used as a probe to screen a cDNA library prepared from flower buds of the apple cultivar (cv.) ‘Fuji’. One positive plaque was obtained after screening ∼100,000 plaques. The cDNA derived from the plaque was designated as MdLHP1a1 (DDBJ accession number, AB290726) after Malus × domestica LHP1. Moreover, we screened about 500,000 plaques from the cDNA library with an MdLHP1a1 probe. A total of seven positive plaques were obtained and then subjected to sequence analysis, revealing that they could be classified into four different cDNAs including MdLHP1a1, three of which were designated as MdLHP1a2, MdLHP1b1, and MdLHP1b2 (DDBJ accession numbers, AB290727, AB290728, and AB290729, respectively). The features of four apple LHP1-like cDNAs are collectively shown in Table 1. The alignment in nucleotide sequence showed that MdLHP1a1 was 99.1 and 90.4% identical to MdLHP1a2 and MdLHP1b1, respectively, and that MdLHP1b1 was 99.1% identical to MdLHP1b2. In comparison with the deduced amino acid sequences of the apple LHP1-like genes, four substitutions were found between MdLHP1a1 and MdLHP1a2 and between MdLHP1b1 and MdLHP1b2. On the other hand, 35 substitutions and five deletions/insertions were found between MdLHP1a1 and MdLHP1b1 (Fig. 1a). These results suggested that a pair of MdLHP1a1 and MdLHP1a2 and a pair of MdLHP1b1 and MdLHP1b2 would be allelic genes. The predicted amino acid sequences of MdLHP1a1 shared lower similarity to those of the HP1 homolog proteins from other plant and animal species [45.1% similarity to SlLHP1 (tomato), 41.6% to AtLHP1 (Arabidopsis), 34.4% to ZmLHP1 (maize), 34.2% to OsLHP1 (rice), 31.3% to DcLHP1 (carrot), 16.7% to DmHP1 (fruit fly), and 14.6% to HsHP1 (human)], whereas the CD (79.6, 98.0, 89.8, 87.8, 87.8, 67.3, and 63.3% similarities, respectively), CSD (64.8, 62.0, 66.2, 69.0, 71.0, 22.5, and 32.4% similarities, respectively), and three nuclear localization signal motifs among them were well conserved, except those of DmHP1 and HsHP1 (Fig. 1a). The phylogenetic analysis indicated that apple LHP1 homologs constitute a small clade and are most similar to tomato SlLHP1 in the HP1 family (Fig. 1b).
Comparison of the predicted amino acid sequences of MdLHP1s and HP1 homologs from plant and animal species. a Schematic alignment of the predicted amino acid sequences of HP1-like proteins from apple (MdLHP1a1, MdLHP1a2, MdLHP1b2, and MdLHP1b2, in this paper), tomato (SlLHP1/LeHP1, AAL25116), Arabidopsis (AtLHP1/TFL2, BAB70689), maize (ZmLHP1, AAM93210), rice (OsLHP1, NP_920664), carrot (DcLHP1, BAA25905), fruit fly (DmHP1, P05205), and homo sapiens (HsHP1, NP_036249). Identical residues are outlined in white against black. Highly conserved regions of the chromo domain (CD), chromo shadow domain (CSD), and putative nuclear localization signal (NLS) are indicated by boxes for CD and CSD and underlines for NLS (Gaudin et al. 2001; Kotake et al. 2003; Zemach et al. 2006). b Phylogenetic tree of the HP1-like proteins. The branch lengths are proportional to the sequence divergence. Numbers along branches are bootstrap values (1,000 replicates). The scale bar represents 0.05 substitutions/site
Genomic organization
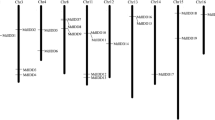
Southern hybridization was performed to assess the copy number of LHP1 homolog genes in the apple genome (Fig. 2). The genomic DNA of the apple cv. ‘Fuji’ was digested with EcoRI, PstI, BamHI, or EcoRV, and the blotted membrane was then subjected to hybridization with full-length MdLHP1a1 cDNA. Two to three major hybridizing bands were detected in each digested sample. In addition, PCR products (4.6 kb) of apple genomic DNA amplified with primers for MdLHP1a1/-a2 were digested with each restriction enzyme used in Southern blot analysis to confirm the internal restriction enzyme sites in the probe region (S1). Furthermore, the 4.6-kb PCR products were cloned to characterize the gene structures of MdLHP1a1/-a2. Eight individual clones were selected randomly, and their sequences were analyzed. Among them, two clones had the same coding sequence as MdLHP1a1, which was designated gMdLHP1a1 (DDBJ accession number, AB292418). The remaining six clones contained the same coding sequence as MdLHP1a2, designated gMdLHP1a2 (DDBJ accession number, AB292419). The gMdLHP1a1 sequence contained PstI and EcoRV sites, whereas gMdLHP1a2 has only an EcoRV site (Fig. 3). The band pattern of the PCR products digested with each restriction enzyme (S1) was consistent with the result of the sequence analysis of them. These findings, together with the results described in the section above, showed that ‘Fuji’ apple had two loci of LHP1 homologs, MdLHP1a and MdLHP1b, each having two alleles, MdLHP1a1/-a2 and MdLHP1b1/-b2, respectively. The MdLHP1a1/-a2 (MdLHP1a) gene comprised six exons and five introns, such as the LHP1 gene (Fig. 3). Moreover, the length of exons was well conserved between LHP1 and MdLHP1a, although the fourth intron in MdLHP1a was about 28 times longer than that in LHP1 (Fig. 3).
Southern hybridization analysis of LHP1 homolog genes in apple. Genomic DNA (10 μg) of ‘Fuji’ apple digested with EcoRI, PstI, BamHI, or EcoRV was electrophoresed in a 0.8% agarose gel, blotted onto nylon membranes, and hybridized with an MdLHP1a1 probe. The molecular size marker is shown in kbp on the left
Schematic representation of the apple MdLHP1a gene in comparison with the Arabidopsis LHP1 gene. The PCR products covering the MdLHP1a1 and MdLHP1a2 alleles are shown as a solid line, and the arrowheads indicate the region for gene-specific primers. Introns are shown as solid lines, while exons are shown as black boxes and numbered with Roman numerals above the black boxes. The lengths (bp) of introns and exons are indicated below (introns) and above (exons) the map. The scale bar represents 500 bp
Expression patterns of MdLHP1a and MdLHP1b in various tissues and in apical buds throughout the growing season in apple
We performed RT-PCR analysis to clarify the expression patterns of MdLHP1a and MdLHP1b in apple. Total RNAs were prepared from various tissues, such as floral buds before bud break in late April; receptacles; apical buds of FBS in August; leaves of the apple cv. ‘Fuji’ in the adult phase; stems, apical buds of VS, and leaves of 1-month-old apple seedlings; and cotyledons and roots of 6-day-old apple seedlings in the juvenile phase. Transcripts of MdLHP1a were detected mainly in apical buds in both the adult and juvenile phases (Fig. 4a). On the other hand, transcripts of MdLHP1b were detected in all tissues used, with relatively higher expression in the apical buds of FBS in the adult phase (Fig. 4a). To investigate the change of the expression of MdLHP1a and MdLHP1b in the apical buds of FBS, which have flowers and fruit in the following year, and SS, which are called water sprouts and have no flowers or fruit, RT-PCR analyses for MdLHP1a and MdLHP1b were carried out using total RNAs extracted from the apical buds of FBS and SS shoots of the apple cv. ‘Jonathan’ from June to October 2004. ‘Jonathan’ apple was used in the seasonal expression analysis, because flower development of this cultivar had been studied well. It is noteworthy that the flowering stage of ‘Jonathan’ apple was early May, and the time of floral bud differentiation was early July in Morioka, Japan (Kotoda et al. 2000). MdLHP1a and MdLHP1b were expressed constantly in apical buds of both kinds of shoots throughout the growing season (Fig. 4b).
Expression analysis of MdLHP1a and MdLHP1b. a RT-PCR products of MdLHP1a and MdLHP1b in various tissues. Total RNAs were isolated from flower buds, fruit receptacles, and apical buds of fruit-bearing shoots (FBS) on August 7, from mature leaves of ‘Fuji’ apple in the adult phase, and from stems, apical buds of vegetative shoots (VS), and leaves of 1-month-old seedlings and cotyledons and roots of 6-day-old seedlings in the juvenile phase. b RT-PCR products of MdLHP1a and MdLHP1b in apical buds of FBS or in apical buds of succulent shoots (SS) of ‘Jonathan’ apple from June to October in 2004. The date of sampling the apical buds of both shoots is indicated above the lanes. Apple Histone H3 was used as an internal control of gene expression
Rescue of the Arabidopsis tfl2 (lhp1) mutant by constitutive expression of apple MdLHP1a1 or MdLHP1b1
In order to analyze the functions of MdLHP1a and MdLHP1b using transgenic Arabidopsis, we generated two chimeric constructs, 35SΩ:MdLHP1a1 and 35SΩ:MdLHP1b1 (Fig. 5a). The infiltration method (Bechtold et al. 1993) was employed to introduce those constructs into the tfl2-2 (lhp1) null mutant (Kotake et al. 2003). Consequently, seventeen and seven independent transgenic lines were obtained for the constructs 35SΩ:MdLHP1a1 and 35SΩ:MdLHP1b1, respectively (described as 35SΩ:MdLHP1a1/tfl2-2 and 35SΩ:MdLHP1b1/tfl2-2 for each transgenic plant, respectively). T3 transgenic plants were grown under LD conditions (16 h light/8 h darkness) for the examination of their phenotypes, such as plant size, inflorescence morphology, and flowering time (the number of rosette leaves produced at flowering). Five lines of 35SΩ:MdLHP1a1/tfl2-2 and four of 35SΩ:MdLHP1b1/tfl2-2 were selected to examine the phenotypes. In the representative transgenic lines of 35SΩ:MdLHP1a1/tfl2-2 (#1) and 35SΩ:MdLHP1b1/tfl2-2 (#2), the phenotypes, such as the small plant size and curled leaves that are characteristic of the tfl2-2 mutant, were rescued (Fig. 5b). Inflorescence shoots, including secondary ones, kept producing flowers in the transgenic lines of 35SΩ:MdLHP1a1/tfl2-2 and 35SΩ:MdLHP1b1/tfl2-2 (Fig. 5e, f), showing that both MdLHP1a1 and MdLHP1b1 rescued the determinate inflorescences of the tfl2-2 mutant. The flowering time of the transgenic plants was slightly earlier than that of the wild-type plants but later than that of the tfl2-2 mutants (Table 2). 35SΩ:MdLHP1a1/tfl2-2 (#1) plants flowered with 8.0 rosette leaves, and 35SΩ:MdLHP1b1/tfl2-2 (#2) plants flowered with 8.8 rosette leaves. In contrast, wild-type plants flowered with 8.9 rosette leaves, and tfl2 plants flowered with 5.3 rosette leaves. The cell size in leaves of the lhp1 (tfl2) mutant is different from that of the wild type under LD conditions (Gaudin et al. 2001). The plant size of wild-type Arabidopsis plants increases more when they are grown under SD conditions (8 h light/16 h darkness) than under LD conditions. Because the tfl2 mutant is photoperiod-insensitive, the cell size of the tfl2 mutant was apparently smaller than that of the wild type under SD conditions (Fig. 5g, h). We observed the upper epidermis of a third rosette leaf in the 35SΩ:MdLHP1a1/tfl2-2 (#1) plant under SD conditions, showing partial, if not complete, restoration in the cell size of the rosette leaves (Fig. 5i).
Constitutive expression of MdLHP1a1 and MdLHP1b1 in the Arabidopsis tfl2-2 mutant and wild-type plants: effects on flowering time and growth. a Schematic representation of the transformation vectors, 35SΩ:MdLHP1a1 and 35SΩ:MdLHP1b1 used in this study. b Seventeen-day-old plants after sowing. From left to right, tfl2-2, wild-type, 35SΩ:MdLHP1a1/tfl2-2 (#1), and 35SΩ:MdLHP1b1/tfl2-2 (#2). c–f Inflorescences of the plants shown in b, which were photographed 23 days after sowing. g–i Micrographs of the epidermal cells on a third rosette leaf from tfl2-2 mutant plants (g), wild-type plants (h), and 35SΩ:MdLHP1a1/tfl2-2 (#1) transgenic plants (i), which were grown on MS medium plates under short-day (SD) conditions (8 h light/16 h dark). j Twenty-three-day-old wild-type plants and 35SΩ:MdLHP1a1/wt (#3) transgenic plants. k Hybridization pattern with an MdLHP1a1 probe for total RNA from transgenic and control Arabidopsis plants. A photograph of the ethidium bromide-stained gel before blotting is shown below. An arrowhead indicates the expected size of the full-length mRNA of MdLHP1a1 and MdLHP1b1. The molecular size marker is shown in kbp on the right. Scale bars are 2 cm in b and j, 5 cm in c–f, and 20 μm in g–i. The plants shown in b, c–f, and j were grown in potted soil under long-day (LD) conditions (16 h light/8 h dark)
To examine whether transgenic Arabidopsis with 35SΩ:MdLHP1a1 or 35SΩ:MdLHP1b1 shows additional novel phenotypes, these constructs were also introduced into the wild-type Arabidopsis (Columbia). More than thirty kanamycin-resistant lines were obtained for each construct (described as 35SΩ:MdLHP1a1/wt or 35SΩ:MdLHP1b1/wt, respectively). Most of the T1 transgenic plants showed slightly delayed flowering (70–80% of all lines), but some of them showed pleiotropic phenotypes, such as the tfl2-2 mutants with early flowering, terminal flowers, and curled leaves (10%) (data not shown). These phenotypes were inherited by subsequent generations. The phenotypes of five representative T3 transgenic lines of 35SΩ:MdLHP1a1/wt and four of 35SΩ:MdLHP1b1/wt showing delayed flowering were examined under LD conditions. 35SΩ:MdLHP1a1/wt (#3) plants flowered with 10.4 rosette leaves, whereas wild-type plants flowered with 8.9 rosette leaves (Fig. 5j; Table 2). Delayed growth of transgenic plants with MdLHP1a or MdLHP1b constructs appeared to develop a few additional rosette leaves, although the appearance of the transgenic plants was similar to that of the wild-type plants. Thus, constitutive expression of MdLHP1a1 or MdLHP1b1 could affect flowering time in the wild-type Arabidopsis plants.
Northern hybridization analysis in transgenic Arabidopsis with MdLHP1a1 or MdLHP1b1
To determine whether or not the expression level of the transgene is correlated with the degree of the phenotype in transgenic plants, Northern hybridization analysis was performed using a DIG-labeled MdLHP1a1 probe. As a result, the expression of the transgenes was confirmed in all transgenic lines with the tfl2-2 mutant background (Fig. 5k). In contrast, several lines showed little expression of the transgenes with the wild-type background (Fig. 5k). The degree of delayed flowering was not exactly due to the expression level of MdLHP1a1 or MdLHP1b1 in transgenic plants, although the delayed flowering in these transgenic plants appeared to depend on the expression of MdLHP1a1 or MdLHP1b1 (Fig. 5k). For example, lines 35SΩ:MdLHP1a1/tfl2-2 #2 and #9 were considerably different in the expression level of MdLHP1a1, but there was little difference between them in the total numbers of rosette leaves produced at flowering (Fig. 5k; Table 2). A similar result was obtained in lines 35SΩ:MdLHP1a1/wt #2 and #5 (Fig. 5k; Table 2). In addition, smaller than expected sizes of the MdLHP1a1/MdLHP1b1 transcripts were detected, predominantly in the transgenic plants with the tfl2-2 background, as compared with those with the wild-type background (Fig. 5k).
Discussion
In this study, we isolated and characterized four cDNAs of apple LHP1 homologs, MdLHP1a1, MdLHP1a2, MdLHP1b1, and MdLHP1b2. The genomic Southern hybridization and sequence analyses revealed that the apple cv. ‘Fuji’ had two loci of LHP1 homologs, MdLHP1a and MdLHP1b, each having two alleles, MdLHP1a1/-a2 and MdFL2b1/-b2, respectively, and that the genomic structure of MdLHP1a was similar to that of Arabidopsis LHP1 (Fig. 3). A comparison of the amino acid sequences revealed that the plant LHP1 homologs exhibited lower similarity with each other, suggesting that a rapid divergence of these genes had occurred among plant species through evolution (Fig. 1a, b). In accordance with the dynamic modification of the chromatin structures in the evolutionary timescale, the LHP1 family regulating the chromatin structure might also have evolved rapidly and been widely distributed among plant species. However, CD, CSD, and nuclear localization signal motifs characteristic of the LHP1 family were highly conserved in apple and other plant LHP1 homologs (Fig. 1a). In animals, the CD of a human PcG is mainly involved in recognition of histone modifications and essential for protein-protein interaction within a multimeric protein complex; the CSD of HP1 protein is involved in self-association and interaction with the chromatin assembly factor 1 (CAF1) and the transcriptional intermediary factor 1β (TIF1β) proteins (Brasher et al. 2000; Eissenberg 2001; Cammas et al. 2004). In plants, the CD of SlLHP1 (LeHP1, tomato LHP1) is required for the interaction of SlLHP1 with K9-methylated Histone H3, which was revealed by Glutathione-S-transferase (GST) pull-down assays in vitro (Zemach et al. 2006). Furthermore, the CSD of both AtLHP1 and SlLHP1 is responsible for homo-dimerization (Gaudin et al. 2001; Zemach et al. 2006) and SlLHP1 CSD could participate in interaction with chromocenters in Arabidopsis cells (Zemach et al. 2006). In this study, we demonstrated that the constitutive expression of MdLHP1a1 or MdLHP1b1 could restore the flowering time and cell size of rosette leaves in the tfl2-2 mutant (Fig. 5; Table 2). Observations of transgenic tfl2-2 plants with MdLHP1a1/-b1 have shown that these genes function equivalently to Arabidopsis LHP1. Based on these results, LHP1 homolog genes in plants are essential for its growth, as they are in animals; CD and CSD, which are highly conserved in HP1 family, would especially be important for the biological function of HP1 homolog proteins.
MdLHP1a and MdLHP1b were constitutively and abundantly expressed in apical buds of proliferating fruit-bearing and succulent (vegetative) shoots of apple trees throughout the growing season (Fig. 4). In transgenic Arabidopsis expressing MdLHP1a1 or MdLHP1b1 both in tfl2-2 and wild-type backgrounds, the amount of the transcripts of MdLHP1a1 and MdLHP1b1 might not be correlated with the extent of the phenotypes, such as flowering time (Fig. 5k; Table 2). In Arabidopsis, LHP1 and EMF2, which maintain the gene activity states epigenetically, and are expressed predominantly in proliferating cells. In addition, overexpression of these genes does not affect the growth phases. These observations suggest that HP1-family and PcG genes are post-transcriptionally regulated (Gaudin et al. 2001; Yoshida et al. 2001; Hsieh et al. 2003; Kotake et al. 2003). It was consistent with these observations that the phenotype of transgenic plants with MdLHP1a1/-b1 was not necessarily correlated with the extent of transcription level. In Northern blot analysis, the aberrant or incomplete transcripts of MdLHP1a1/-b1 were found in transgenic Arabidopsis plants with MdLHP1a1/-b1, predominantly in the tfl2-2 mutant background. On the other hand, few aberrant transcripts were found in transgenic plants with MdLHP1a1/-b1 in the wild-type background, although the expression of MdLHP1a1/-b1 in the wild-type background was no less than that in the tfl2-2 background. The lower level of aberrant transcripts in the wild-type background may be due to a mechanism that represses the abnormal accumulations of the functional transcripts of LHP1-like genes. In some (10%) of the transgenic Arabidopsis with 35SΩ:MdLHP1a1/-b1 in the wild-type background, down-regulation or silencing of endogenous LHP1 might cause the expression of tfl2-2 mutant-like phenotypes by a mechanism, such as co-suppression or RNA interference (RNAi).
In addition to MdLHP1a and MdLHP1b, several pairs of two duplicated genes regulating floral identity or flowering time have been identified in the apple genome; for example, pairs of APETALA1 (AP1)-like genes from the apple cv. ‘Granny Smith’, ‘Jonathan’, ‘Fuji’, and ‘Pinova’ (Yao et al. 1999; Kotoda et al. 2000, 2002; our unpublished results), LEAFY (LFY)-like genes from ‘Jonathan’ and ‘Fuji’ (Wada et al. 2002; Esumi et al. 2005), and TFL1-like genes from ‘Jonathan’ and ‘Fuji’ (Esumi et al. 2005; Kotoda and Wada 2005; Kotoda et al. 2006; our unpublished results) have been isolated and characterized. The pair of paralogous genes related to apple flowering seems to have similar regulatory functions, considering the results from the analysis of transgenic Arabidopsis ectopically expressing those genes, although the expression levels and patterns in apple are not identical between the twin paralogs. The fact that there are pairs of two copy genes in the apple genome supports the hypothesis that the Maloideae subfamily (a basal haploid number, n = 17), which includes the genera Malus (apple) and Pyrus (pear), had evolved from the polyploid ancestor. It was hypothesized that Maloideae subfamily had been created by hybridization between ancestors of two other Rosaceae subfamilies, Spiraeoideae (n = 9) and Amygdaloideae (n = 8) (Sax 1933; Luby 2003) or that the origin of Maloideae (n = 18) had occurred from members of a lineage containing the ancestors of Gillenia (n = 9), and then its progeny had lost one chromosome (n = 17) (Evans and Campbell 2002). Because polyploidy (genome doubling) can easily produce functional divergence among homolog genes, this is one of the evolutional strategies in plants that enhances environmental adaptation (Adams and Wendel 2005). As an example of studies on such an evolutionary polyploid complex genome, the chromosomal rearrangement of amphidiploid Brassica napus (AACC) resynthesized by hybridization between allogenic B. rapa (AA) and B. oleracea (CC) affects the gene expression of FLOWERING LOCUS C (FLC) homologs, leading to a divergence in flowering time (Pires et al. 2004; Adams and Wendel 2005). In consideration for Brassica genome study, the polyploid complex genome may produce a variety of phenotypes in apple as well. Sung et al. (2006) reported that Arabidopsis LHP1 also plays a crucial role in vernalization by regulating FLC. Interestingly, the vernalization requirement in some Arabidopsis ecotypes for its development resembles the chilling requirement in deciduous trees, such as the apple, which are adapted to survive at cooler temperatures in winter, although the apple is a perennial plant and its life cycle is different from that of annual plants, such as Arabidopsis.
Taken together, LHP1 homolog genes of chromatin regulator might have rapidly evolved and be diverged widely among plant species following the dynamic modification of chromatin structures with chromosomal rearrangement during evolution, but the protein functions were conserved, at least between Arabidopsis and apple. Recently, we produced transgenic apples with an RNAi construct for MdLHP1a/MdLHP1b. These transgenic approaches may provide not only the insight into the practical aspects, such as the manipulation of flowering and the acceleration of breeding, but also a novel notion of the mechanism controlling flowering or dormancy via epigenetic system in fruit trees.
Abbreviations
- FBS:
-
Fruit-bearing shoot
- LD:
-
Long-day
- SD:
-
Short-day
- SS:
-
Succulent shoot
- VS:
-
Vegetative shoot
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316:1194–1199
Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J 19:1587–1597
Cammas F, Herzog M, Lerouge T, Chambon P, Losson R (2004) Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev 18:2147–2160
Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J (2004) Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131:5263–5276
Dennis F Jr (2003) Flowering, pollination and fruit set and development. In: Ferree DC, Warrington IJ (eds) Apples: botany, production and uses. CAB Intl, Oxon, Wallingford
Eissenberg JC (2001) Decisive factors: a transcription activator can overcome heterochromatin silencing. Bioessays 23:767–771
Esumi T, Tao R, Yonemori K (2005) Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sex Plant Reprod 17:277–287
Evans RC, Campbell CS (2002) The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. Am J Bot 89:1478–1484
Gallie DR, Walbot V (1992) Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res 20:4631–4638
Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O (2001) Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128:4847–4858
Germann S, Juul-Jensen T, Letarnec B, Gaudin V (2006) DamID, a new tool for studying plant chromatin profiling in vivo, and its use to identify putative LHP1 target loci. Plant J 48:153–163
Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386:44–51
Hackett WP (1985) Juvenility, maturation, and rejuvenility in woody plants. Hortic Rev 7:109–155
Hood EE, Helmer GL, Fraley RT, Chilton MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168:1291–1301
Hsieh TF, Hakim O, Ohad N, Fischer RL (2003) From flour to flower: how Polycomb group proteins influence multiple aspects of plant development. Trends Plant Sci 8:439–445
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405
Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K (2003) Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol 44:555–564
Kotoda N, Iwanami H, Takahashi S, Abe K (2006) Antisense expression of MdTFL1, a TFL1-like gene, reduces the juvenile phase in apple. J Am Soc Hortic Sci 131:74–81
Kotoda N, Wada M (2005) MdTFL1, a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis. Plant Sci 168:95–104
Kotoda N, Wada M, Komori S, Kidou S, Abe K, Masuda T, Soejima J (2000) Expression pattern of homologues of floral meristem identity genes LFY and AP1 during flower development in apple. J Am Soc Hortic Sci 125:398–403
Kotoda N, Wada M, Kusaba S, Kano-Murakami Y, Masuda T, Soejima J (2002) Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci 162:679–687
Larsson AS, Landberg K, Meeks-Wagner DR (1998) The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149:597–605
Luby JJ (2003) Taxonomic classification and brief history. In: Ferree DC, Warrington IJ (eds) Apples: botany, production and uses. CAB Intl, Oxon, Wallingford
Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5:296–304
Minc E, Courvalin JC, Buendia B (2000) HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet 90:279–284
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Nakahigashi K, Jasencakova Z, Schubert I, Goto K (2005) The Arabidopsis HETEROCHROMATIN PROTEIN1 Homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol 46:1747–1756
Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation, EMBnet News 4:1–4
Perrière G, Gouy M (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369
Pires JC, Zhao JW, Schranz ME, Leon EJ, Quijada PA, Lukens LN, Osborn TC (2004) Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol J Linn Soc Lond 82:675–688
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Sax K (1933) The origin of the Pomoideae. Proc Am Soc Hortic Sci 30:147–150
Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38:706–710
Takada S, Goto K (2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15:2856–2865
Wada M, Cao QF, Kotoda N, Soejima J, Masuda T (2002) Apple has two orthologues of FLORICAULA/LEAFY involved in flowering. Plant Mol Biol 49:567–577
Yao JL, Dong YH, Kvarnheden A, Morris B (1999) Seven MADS-box genes in apple are expressed in different parts of the fruit. J Am Soc Hortic Sci 124:8–13
Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S (2001) EMBRYONIC FLOWER2, a novel Polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13:2471–2481
Zemach A, Li Y, Ben-Meir H, Oliva M, Mosquna A, Kiss V, Avivi Y, Ohad N, Grafi G (2006) Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei. Plant Cell 18:133–145
Acknowledgments
We thank Dr. H. Ichikawa, National Institute of Agrobiological Sciences (Tsukuba, Japan), for kindly providing the binary vectors, pSMAK312Blue and pSMAK193E. We also thank Dr. E.E. Hood, Texas A&M University College Station, for kindly providing Agrobacterium tumefaciens EHA101. A. thaliana tfl2-2 seeds were kindly provided by the Arabidopsis Biological Resource Center at Ohio State University. This work was supported by the “PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences)” from the Bio-Oriented Technology Research Advancement Institution (BRAIN). This publication constitutes the contribution number 1445 of the National Institute of Fruit Tree Science.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2007_250_MOESM1_ESM.pdf
S1. Restriction fragments of PCR products of the MdLHP1a gene PCR products of the MdLHP1a gene corresponding to the probe region used in Southern blot analysis were digested with EcoRI, PstI, BamHI, or EcoRV. The molecular size marker is shown in kbp on the left. (PDF 69.6 kb)
Rights and permissions
About this article
Cite this article
Mimida, N., Kidou, Si. & Kotoda, N. Constitutive expression of two apple (Malus × domestica Borkh.) homolog genes of LIKE HETEROCHROMATIN PROTEIN1 affects flowering time and whole-plant growth in transgenic Arabidopsis . Mol Genet Genomics 278, 295–305 (2007). https://doi.org/10.1007/s00438-007-0250-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0250-0