Abstract
Chicken coccidiosis is caused by the apicomplexan parasite Eimeria spp. At present, drug resistance of Eimeria is common because of the indiscriminate use of anticoccidial drugs. The gene encoding surface antigen 10 of Eimeria tenella (EtSAG10) is differentially expressed between drug-resistant and drug-sensitive strains. RNA-seq analysis indicated that this gene was downregulated in strains resistant to maduramicin and diclazuril compared to susceptible strains. EtSAG10 DNA sequence alignment revealed that they contained one and ten mutations in MRR and DZR, compared with DS, respectively. A full-length EtSAG10 cDNA was successfully cloned and expressed, and the polyclonal antibody was prepared. The transcription and translation levels of EtSAG10 were analyzed by quantitative real-time PCR (qPCR) and Western blotting. The localization of EtSAG10 in Spz, Mrz, and parasites in the first asexual stage was determined by indirect immunofluorescence. The potential association of EtSAG10 with sporozoite invasion of host cells was assessed by invasion inhibition assays. The results showed that EtSAG10 had a predicted transmembrane domain at the C-terminal end and a predicted signal peptide at the N-terminal end. EtSAG10 was downregulated in drug-resistant strains, which is consistent with the RNA-seq results. The EtSAG10 protein was localized to the parasite surface and parasitophorous vacuole membrane. This protein was shown to play a role in the infection of chicken intestine by sporozoites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chicken coccidiosis is a parasitic disease caused by the apicomplexan protozoan Eimeria spp. and causes considerable economic losses to the poultry industry worldwide. Eimeria tenella parasitizes chicken cecal epithelial cells and is one of the most pathogenic species among the seven identified species (Williams 2002). The current control strategies for coccidiosis rely on anticoccidial drugs and live oocyst vaccines (Sun et al. 2014a; Shirley et al. 2005).
However, the extensive use of anticoccidial drugs has resulted in the development of resistance in Eimeria spp. And the use of these drugs has been restricted by the emergence of resistance (Chapman 1997). Eimeria spp. have been shown to be resistant to all available anticoccidial agents. A Dutch study evaluated various E. tenella strains and found that their sensitivity to drugs was reduced over time (Peek and Landman 2003). Several studies investigated drug resistance in Eimeria (Jiang et al. 2005; Chen et al. 2008; Thabet et al. 2017). Nonetheless, the underlying molecular mechanisms of resistance are not fully understood. Therefore, exploring the function of resistance-related genes is essential.
Our research group previously used the RNA-seq method to compare differences between maduramicin- and diclazuril-resistant strains and drug-sensitive strains of E. tenella and screened for differentially expressed genes and found that the expression of surface antigen 10 of E. tenella (EtSAG10) was downregulated in two resistant strains (data unpublished). Transcriptome sequencing showed that the log2Ratio (DZR/DS) was reduced by − 1.5 and the log2Ratio (MRR/DS) was reduced by − 2.7.
Therefore, the role of EtSAG10 in drug-resistant strains deserves further exploration. More than 80 SAGs are expressed in E. tenella. EtSAG10 is one of the surface antigen (SAG) subfamily members; all known subfamilies encode signal peptides and addition sites for GPI anchors (Reid et al. 2014). Some SAGs induce pro-inflammatory cytokine responses in E. tenella in vitro (Chow et al. 2011). Moreover, SAG genes in E. tenella encode single-domain, membrane-bound proteins tethered by GPI anchors to the surface of invasive sporozoites and merozoites (Tabares et al. 2004). E. tenella SAG1 binds to mammalian cells (Jahn et al. 2009). Several SAGA subfamily proteins, but not SAGB subfamily proteins, can bind to cultured cells (Reid et al. 2014). The surface antigens of other apicomplexan protozoans such as Toxoplasma gondii (T. gondii) can be potentially used as diagnostic antigens and vaccines (Leng et al. 2014). TgSAG1 elicited protective responses to T. gondii infection (Aosai et al. 1999), and TgSAGs can stimulate host innate and adaptive immune responses through Toll-like receptors (Gazzinelli and Denkers 2006). In addition, SAGs are believed to play an important role in the early recognition of protozoa, adherence to and invasion of host cells, immune regulation, and immune escape )Cardwell and Martinez 2009; Gould et al. 2017; Leal-Sena et al. 2018). The number of studies on SAGs has increased in recent years.
EtSAG10 had not been characterized in E. tenella to date. In this study, EtSAG10 DNA sequences of three strains were aligned by using the Clustal X tool, and the full-length EtSAG10 cDNA was cloned and expressed, and its biological functions were determined. The differential expression of SAG10 in drug-resistant and drug-sensitive strains of E. tenella was evaluated. This study will lay the foundation for identifying molecular markers of drug resistance.
Materials and methods
Animals, parasites, and cells
Day-old yellow-feathered broiler chickens were purchased from the Shanghai Fengxian District, China, and were reared in a coccidia-free animal facility. New Zealand rabbits were obtained from the Songlian Experimental Animal Farm. BALB/c mice were purchased from the Slack Laboratory Animal Limited Company.
The Shanghai drug-sensitive (DS) strain of E. tenella was collected and isolated in a chicken farm in Shanghai, China, in the 1980s, and was kept in our laboratory (Huang et al. 1993). A maduramicin-resistant strain (MRR) and diclazuril-resistant strain (DZR) were induced and maintained in our laboratory (Han et al. 2004). Parasites were propagated by passage through coccidia-free 2-week-old chickens, as described previously, and stored in 2.5% potassium dichromate solution (Tomley 1997). Sporozoites (Spz) were obtained from sporulated oocysts, and unsporulated oocysts (UO) and sporulated oocysts (SO) were obtained as previously described (Shirley 1995; Han et al. 2010). Second-generation merozoites (Mrz) were collected and purified using Percoll density gradient centrifugation (Zhou et al. 2010).
The chicken embryo fibroblast cell line DF-1 was kept in our laboratory and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, USA) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, USA), 100 U/mL penicillin/streptomycin (Invitrogen), and 2.0 mM l-glutamine (Invitrogen).
Analysis of the DNA sequence of EtSAG10 in three strains
Genomic DNA was extracted from sporulated oocysts of two drug-resistant strains and the drug-sensitive strain of E. tenella according to the instructions of TIANamp Genomic DNA Kit (Tiangen, China), respectively. The DNA sequence of EtSAG10 (ID: ETH_00034975) was amplified using the following primers: 5′-ATGCTGCAGCGGAAGCTACCACCCA-3′, 5′-TCATAAAGTCATAATGCCGAACGTC-3′. The 1168-bp amplified DNA sequences were cloned into the pGEM-T easy cloning vector (Promega, Madison, WI, USA) and sent to Sangon Biotech (China) for sequencing. Further, the DNA sequences of the three strains were aligned by using Clustal X tool. The DNA sequences were sequenced and aligned in triplicate.
RNA extraction, cDNA synthesis, and gene cloning
Total RNA was extracted from sporulated oocysts of a drug-sensitive strain of E. tenella using Trizol (Invitrogen). The concentration and purity of RNA were determined by measuring the optical density at 260 and 280 nm using an ultraviolet spectrophotometer (Eppendorf, Hamburg, Germany). The RNA was treated with DNase I (Invitrogen) and reverse transcribed into cDNA using a SuperScript™ III reverse transcriptase kit (Invitrogen) and Oligo (dT) primers.
The full-length coding region of the EtSAG10 gene (ID: ETH_00034975) was amplified from the cDNA of E. tenella using the following primers with the BamHI and SalI restriction sites (underlined): 5′-GCGGATCC ATGCTGCAGCGGAAGCTACCACCCA-3′, 5′-GCGTCGACTCATAAAGTCATAATGCCGAACGTC-3′. The 786-bp amplified DNA fragment was cloned into the pGEM-T easy cloning vector (Promega, Madison, WI, USA) and transformed into Escherichia coli Top10. The bacteria transformed with recombinant plasmid pGEM-T-EtSAG10 was sent to Sangon Biotech (China) for sequencing. The gene sequences and deduced amino acid sequences of EtSAG10 were analyzed using BLAST programs from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), the genome sequence of E. tenella (http://www.genedb.org/Homepage/Etenella), translation tools from the ExPASy server (http://www.expasy.org/tools/protparam.html), a prediction tool for membrane protein topology and signal peptides (http://octopus.cbr.su.se/index.php), and Motif Scan (https://myhits.isb-sib.ch/cgi-bin/motif_scan) (Zhai et al. 2016).
Protein expression and purification
The ORF was excised from recombinant pGEM-T-EtSAG10 plasmids by digestion with Bam HI and Sal I after sequencing and ligated into the prokaryotic expression vector pGEX-4T-2 (Novagen, Germany) digested with the same restriction endonucleases. Recombinant expressed plasmids were extracted from the clones and subjected to DNA sequencing to confirm the sequence and orientation of the inserts. The pGEX-4T-EtSAG10 plasmids were expressed using an E. coli BL21 (DE3) expression system (Tiangen). Bacteria carrying pGEX-4T-EtSAG10 were grown in 200 mL Luria Broth medium containing 50 mg/mL at 37 °C. When the culture growth reached an optical density of 0.6 at 600 nm, protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich, St. Louis, USA) at 37 °C for 8 h. The recombinant bacteria were isolated by centrifugation at 10,000g for 15 min, and the pellet was then suspended in 20 mL phosphate-buffered saline (PBS, pH 7.4) and extracted using an ultrasonic processor to release the fusion proteins. Lysates were centrifuged at 10,000g for 10 min to separate inclusion bodies and cellular debris from the remaining soluble substances. The pellet was resuspended in 5 mL 1× binding buffer plus 6 M urea. Analysis by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) demonstrated that recombinant EtSAG10 (rEtSAG10) was expressed in soluble form. The soluble rEtSAG10 was purified by using GST-bind resin (Merck, Darmstadt, Germany) according to the manufacturer’s instructions. The purified protein was separated on 12% SDS-PAGE and stained with Coomassie Blue. Protein concentration was determined using a BCA protein assay kit (Beyotime, Haimen, China).
Preparation of polyclonal antibodies against EtSAG10
Two-month-old rabbits and 6-week-old mice were immunized with 0.2 mg and 0.05 mg of purified rEtSAG10 per animal, respectively. rEtSAG10 was emulsified with the same volume of Freund’s complete adjuvant (Sigma-Aldrich) used at the immunization. After that, experimental animals were injected with rEtSAG10 emulsified with the same volume of Freund’s incomplete adjuvant (Sigma-Aldrich). After three immunizations, the sera of rabbits and mice were collected. Reactogenicity of rEtSAG10 was determined by Western blotting.
Quantitative real-time PCR (qPCR)
The expression profile of EtSAG10 mRNA was examined at different developmental stages (UO, SO, Spz, and Mrz) in the DS strain of E. tenella and in SO of the strains DS, DZR, and MRR using qPCR. Total RNAs were isolated using TRIzol reagent (Invitrogen, USA) following the manufacturer’s instructions. All samples were treated with deoxyribonuclease I (Invitrogen). Total RNAs (35 μg/reaction) were reverse transcribed into cDNA using the SuperScript™ III Reverse Transcriptase kit (Invitrogen) and random primers, respectively. The housekeeping gene 18S rRNA was used as an internal control (Livak and Schmittgen 2001; Kumar et al. 2012; Wang et al. 2016). The qPCR primers for EtSAG10 were 5′-TCCAGCACCAGAGGAAGGAGAACTA-3′ (sense) and 5′-TGAAGGCGGGAGTCCAGTGAAGTTT-3′ (antisense). The qPCR primers for 18S rRNA were 5′-TGTAGTGGAGTCTTGGTGATTC-3′ (sense) and 5′-CCTGCTGCCTTCCTTAGATG-3′ (antisense). qPCR was performed using the SYBR® Premix DimerEraser™ (Perfect Real Time) (TaKaRa, Japan) kit in a QuantStudio®5 Real-Time PCR system. The 2−∆∆Ct method (Geysen et al. 1991; Livak and Schmittgen 2001) was applied for measuring relative changes in gene expression.
Western blotting
Parasites at four developmental stages (UO, SO, Spz, and Mrz) of E. tenella DS strain and parasites at the SO stage in strains DS, DZR, and MRR were lysed in RIPA buffer (Beyotime) and Protease Inhibitor Cocktail (Sigma) and centrifuged at 4000g for 10 min. The supernatants or purified rEtSAG10 were incubated with SDS sample buffer and denatured by heating. The protein samples were separated on a 12% SDS-PAGE gel electrophoresis and blotted onto Immobilon-P transfer membranes (Millipore). The membranes were blocked with phosphate-buffered saline (PBS) containing 5% skimmed milk powder for 2 h at 37 °C, washed three times with PBS for 5 min, and incubated with anti-α-tubulin monoclonal antibody (1:400 dilution), GST-monoclonal antibody, polyclonal anti-rEtSAG10 (1:200 dilution) and anti-sporozoite rabbit serum (1:200 dilution) which observed in our lab (Han et al. 2015) for 2 h at 37 °C. After that, the membranes were washed three times with PBST for 5 min and incubated with IRDye® 680RD donkey anti-mouse IgG and IRDye® 800CW goat anti-rabbit antibody IgG (LI-COR Biosciences, USA, 1:10,000) diluted in PBS for 1 h at 37 °C in the dark and visualized using an Odyssey® Infrared Imaging System (LI-COR Biosciences). Alpha-tubulin (Sigma) was used as a reference protein for normalization.
Localization of EtSAG10 by immunofluorescence microscopy
The immunolocalization of EtSAG10 at different developmental stages of E. tenella was performed as described previously (Peroval et al. 2006; Jiang et al. 2012). To confirm the location of EtSAG10 in the parasite, purified sporozoites and second-generation merozoites were transferred to glass slides and air-dried. The chicken embryo fibroblast cell line DF-1 was used for in vitro infection experiments. The cells were cultured in six-well plates (Corning, NY, USA) with Φ25mm Circle Microsope Cover Glass (NEST) and cultured in complete medium [DMEM (Invitrogen) containing 10% FBS, 100 U/mL penicillin/streptomycin] at 37 °C and 5% CO2 for 10 h. Freshly purified sporozoites were incubated in complete medium for 2 h at 41 °C and were added to the cultured cells. At the indicated post-inoculation time points, infected DF-1 cells were fixed, washed, and transferred to glass slides. The slides were fixed in 2% paraformaldehyde in PBS, air-dried, and permeabilized in 1% Triton X-100 in PBS for 15 min. The slides were blocked with 2% (w/v) bovine serum albumin in PBS overnight at 4 °C and incubated with polyclonal anti-rEtSAG10 diluted 1:100 in PBS for 1 h at 37 °C. The slides were incubated with a 1:500 dilution of goat anti-rabbit IgG fluorescein isothiocyanate-conjugated antibody (Sigma-Aldrich) for 1 h at 37 °C. Nuclei were stained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (Beyotime) (10 mg/mL) for 30 min at room temperature. After each step, the slides were washed three times for 10 min with PBS containing 0.05% Tween 20. After that, the slides were mounted using 60 μL Fluoromount Aqueous Mounting Medium (Sigma-Aldrich) and observed under a laser scanning confocal microscope (ZEISS, Germany) and a fluorescence microscope (OLYMPUS, Japan).
Invasion inhibition assays
Invasion inhibition assays were carried out as described previously (Jiang et al. 2012; Wilson et al. 2015). DF-1 cells (2 × 105 cells per well) were cultivated in 24-well plates in complete medium for 12 h at 37 °C and 5% CO2. Freshly purified sporozoites were labeled using the dye carboxyfluorescein diacetate succinimidyl ester (CFSE; Beyotime Biotechnology), according to the manufacturer’s instructions and resuspended in DMEM containing 5% FBS and 500 U/mL penicillin/streptomycin and preincubated with 50 μg/mL, 100 μg/mL, 200 μg/mL, or 400 μg/mL purified anti-rEtSAG10 IgG for 2 h at 37 °C. Sporozoites (6.0 × 105 per well) were added to infected DF-1 cells for 12 h at 41 °C and 5% CO2. The same concentration of rabbit IgG (Sigma-Aldrich, USA) was used as the negative control, and sporozoites incubated without antibodies were used as the positive control. Cells were washed, trypsinized, collected, and detected using a Cytomics FC500 flow cytometer (Beckman Coulter, USA). All assays were performed in triplicate.
Statistical analysis
The relative expression ratio of a target gene was calculated from the PCR efficiency and Ct of the treated sample versus the untreated sample using the ∆∆Ct method. The 18S rRNA gene was used as a reference for normalization. Native rabbit IgG was used as a control in invasion inhibition assays. Statistical analyses were performed using SPSS software version 22.0 (SPSS). Differences between the groups were calculated by one-way analysis of variance, followed by Duncan’s multiple range tests, and the differences were considered statistically significant at P < 0.05.
Ethics statements
The animals were used according to the protocol approved by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Results
EtSAG10 DNA sequence analysis of three strains
The 1168-bp full-length DNA sequences of EtSAG10 were obtained from three strains (DS, DZR, and MRR) of E. tenella, respectively, which included three introns (111 bp to 293 bp, 445 bp to 522 bp, and 899 bp to 994 bp). Three DNA sequences of EtSAG10 of three strains were compared by Clustal X tool (Fig. S1). The results showed that ten point mutations were observed in the EtSAG10 DNA sequence of DZR at the 247th, 660th, 684th, 707th, 712th, 716th, 721st, 757th, 833th, and 946th position and one point mutation was recorded in the EtSAG10 DNA sequence of MRR at the 591th position, compared with DS (Fig. 1). One synonymous mutation in the EtSAG10 DNA sequence, CGA to CGC coding for arginine (Arg) amino acid residue at the 591st position, was found in MRR. Three synonymous mutations in the EtSAG10 DNA sequence was found in DZR, ACG to ACA coding for threonine (Thr) amino acid residue at the 684th position, CAA to CAG coding for glutamine (Gln) amino acid residue at the 660th position, and TTG to CTG coding for leucine (Leu) amino acid residue at the 757th position. Moreover, five missense mutations in the EtSAG10 DNA sequence were found in DZR, CTT to CAT coding for leucine (Leu) amino acid residue changed into histidine (His) amino acid residue at the 707th position, AGA to GGA coding for arginine (Arg) amino acid residue changed into glycine (Gly) amino acid residue at the 712th position, AGG to AAG coding for arginine (Arg) amino acid residue changed into lysine (Lys) amino acid residue at the 716th position, GTG to ATG coding for valine (Val) amino acid residue changed into methionine (Met) amino acid residue at the 721st position, and CCA to CTA coding for proline (Pro) amino acid residue changed into leucine (Leu) amino acid residue at the 833th position.
Sequence analysis of EtSAG10 cDNA
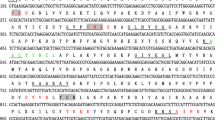
The full-length coding sequence of the EtSAG10 gene in the DS strain of E. tenella was amplified, cloned, and sequenced, and a 786-bp product was obtained. This sequence presented 99% similarity with EtSAG10 previously isolated from E. tenella (GenBank: XM_013375905.1). The newly identified gene encodes a putative protein of 261 amino acid residues with a predicted molecular weight of 27.9 kDa, a theoretical isoelectric point 4.7, a predicted transmembrane domain at the C-terminal end, and a predicted signal peptide at the N-terminal end. The analysis of the predicted motif structure indicated that EtSAG10 contained two casein kinase II phosphorylation sites (residues 143–146 and 149–152), three N-myristoylation sites (residues 61–66, 139–144, and 197–202), three protein kinase C phosphorylation sites (residues 80–82, 143–145, and 201–203), three N-glycosylation sites (residues 136–139, 156–159, and 182–185), and one threonine-rich domain (residues 177–202) (Fig. S2).
Expression and purification of rEtSAG10
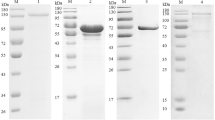
E. tenella SAG10 cDNA was amplified and cloned into a pGEM-T easy cloning vector, subcloned into the expression vector pGEX-4T-2, and expressed in E. coli BL21 (DE3). The bacterial culture was incubated at 37 °C and induced with 0.5 mM IPTG. The SDS-PAGE results demonstrated that the rEtSAG10 was expressed as a soluble protein (Fig. 2a). The experimental size of rEtSAG10 agreed with the predicted size above 53.9 kDa (including 26 kDa the GST tag protein). The expressed protein was purified by immobilized nickel affinity chromatography under native conditions. The Western blot results indicated that the purified protein was recognized by the anti-sporozoite rabbit serum and the GST-monoclonal antibody. Native rabbit IgG failed to detect any protein of the expected size of rEtSAG10. These results indicated that rEtSAG10 has good reactogenicity and that protein rEtSAG10 had been degraded into peptides (Fig. 2b).
a The solubility analysis rEtSAG10, b Western blot analysis of purified rEtSAG10. Rabbit antiserum against E. tenella sporozoite or monoclonal anti-GST antibody was used as the primary antibody. P, pellet; S, supernatant. Lane 1, rEtSAG10 was probed with the naive rabbit serum. Lane 2, rEtSAG10 was probed with the monoclonal anti-GST antibody. Lane 3, rEtSAG10 was probed with the anti-sporozoite rabbit serum
Transcription and translation of EtSAG10 in three E. tenella strains
The mRNA levels of EtSAG10 were measured in the DS, DZR, and MRR strains by qPCR. mRNA transcription in the DZR and MRR strains was lower than that in the DS strain (Fig. 3a). The presence of EtSAG10 in the DS, DZR, and MRR strains was determined by immunoblotting using rabbit antiserum against rEtSAG10. Western blot analysis showed that the protein in the DZR and MRR strains was downregulated compared with that of the DS strain (Fig. 3b, c), and these results agree with the RNA-seq results.
Transcription and translation levels of EtSAG10. DS, drug-sensitive strain; DZR, diclazuril-resistant strain; MRR, maduramicin-resistant strain; UO, unsporulated oocysts; SO, sporulated oocysts; Spz, sporozoites; Mrz, second-generation merozoites. Anti-α-tubulin antibody was used as a loading control. EtSAG10 was recognized by rabbit anti-rEtSAG10. a Transcription level of EtSAG10 in DS, DZR, and MRR. b and cEtSAG10 translation level in DS, DZR, and MRR. d mRNA level of EtSAG10 at different developmental stages in the DS strain of E. tenella. e and fEtSAG10 protein expression level at different developmental stages of the DS strain of E. tenella. The data represent the mean ± S.D. of triplicate determinations and are representative of three independent experiments
Transcription and translation levels of EtSAG10 at different developmental stages of the DS strain
The mRNA expression profile of EtSAG10 was examined in UO, SO, Spz, and Mrz in the DS strain by qPCR. The EtSAG10 mRNA level in SO was higher than that at the other three stages (Fig. 3d). Moreover, the presence of EtSAG10 at four developmental stages in the drug-sensitive strain was determined by immunoblotting using rabbit antiserum against rEtSAG10. Western blotting detected proteins in SO (two bands of approximately 27.9 kDa and greater than 34 kDa), Spz (two bands of approximately 27.9 kDa and greater than 34 kDa), Mrz (two bands of approximately 27.9 kDa and 34 kDa), and UO (one band of approximately 27.9 kDa). Protein expression was higher in SO and UO than in Spz and Mrz (Fig. 3e, f).
Immunofluorescence localization of EtSAG10 in E. tenella-infected DF-1 cells, merozoites, and sporozoites
The localization and distribution of EtSAG10 protein in Spz, Mrz, and parasites in the first asexual stage were evaluated by immunofluorescence. EtSAG10 was predominantly localized to the surface of Spz and Mrz, and green fluorescence intensity on the surface of Mrz was higher than that on the surface of Spz (Fig. 4a, b). After infection of DF-1 cells by sporozoites for 24 h, EtSAG10 was mainly localized to the surface of trophozoites, and green fluorescence intensity was also enhanced at this stage (Fig. 4c). As parasite development progressed in the cells, EtSAG10 was localized to the cytoplasm of immature schizonts, and the intensity of EtSAG10 staining was increased (Fig. 4d–f). EtSAG10 was also detected in the parasitophorous vacuole membrane (PVM) at 60 h (Fig. 4d).
Indirect immunofluorescence to localize EtSAG10 at different developmental stages of Eimeria tenella using rabbit sera against rEtSAG10. a Sporozoites (Spz) in PBS; b second-generation merozoites (Mrz) from infected chicken caeca in PBS; c trophozoites (Tropho) at 24-h post-infection (pi); d immature schizonts at 60 h pi; e immature schizonts at 72 h pi; f immature schizonts at 84 h pi
Inhibition of E. tenella sporozoite invasion by rEtSAG10 polyclonal antibodies
The degree with which the rabbit anti-rEtSAG10 inhibited the invasion of DF-1 cells by E. tenella sporozoites was determined using invasion inhibition assays. Native rabbit IgG was used as a control. Under experimental conditions, the inhibition rate appeared to increase as the antibody concentration increased. The invasion rate was approximately 60% after pretreatment with 400 μg/mL of the anti-rEtSAG10 polyclonal antibody. In contrast, invasion was not significantly inhibited by native rabbit IgG (Fig. 5).
Discussion
At present, Eimeria spp. resistance to drugs is widespread; therefore, it is crucial to quickly diagnose resistance in parasites. In this study, we compared the DNA sequences of EtSAG10 of three strains (DZR, MRR, and DS). Sequence alignment found that they contained missense mutations and synonymous mutations in DZR and MRR, compared with DS. Previous reports showed that mutations in some genes determining resistance to drugs such as artemisinin and antifolates have been identified in Plasmodium (Gregson and Plowe 2005; Mbengue et al. 2015). And it was reported that the mutations were associated with mRNA and protein expression levels (Jia and Zhao 2017). So we speculated that the mutations in two drug-resistant strains may be associated with mRNA and protein expression levels and contribute to the resistance to these two drugs. Moreover, we compared the expression of the EtSAG10 gene between drug-resistant and sensitive strains using different methods. qPCR and Western blot analysis demonstrated that the expression of EtSAG10 in two resistant strains was downregulated compared with susceptible strains. These results agree with previous findings, wherein surface antigens were downregulated in E. tenella MRR (Chen et al. 2008). Similarly, the expression of surface antigens in drug-resistant strains of Plasmodium falciparum and T. gondii was lower than that in sensitive strains (Doliwa et al. 2013; Antony et al. 2016). The results of these studies and the present study evidenced that drug resistance in Eimeria spp. may be related to the downregulation of the expression of SAGs. Therefore, we speculated that the downregulation of EtSAG10 was essential for drug resistance in E. tenella. A previous study found that the downregulation of the surface antigen CD53 in human neutrophils was the result of the activation of a proteolytic mechanism (Mollinedo et al. 1998), which is consistent with our hypothesis.
The function of EtSAG10 was further investigated by determining its expression at different developmental stages of E. tenella by qPCR. EtSAG10 mRNA expression was higher in SO than the other three stages, and these results agreed with Western blotting results. We hypothesized that this result was related to changes in the environment of SO. For instance, it has been reported that the expression of sHSPs is increased under different stress conditions, including heat, cold, chemical intoxication, and nutritional stress (Montero et al. 2008; Perez-Morales et al. 2009; Liu et al. 2012; Sun et al. 2014b). Nonetheless, compared with qPCR results, Western blot results also showed that EtSAG10 protein expression was higher in UO and Mrz; we speculated that the time and location of transcription and translation of eukaryotic genes may be separated by time and space and that the mRNA might have been degraded when the protein levels peaked at different time points of detection (Ben-Ari et al. 2010). In addition, Western blot analysis revealed two bands in SO, Spz, and Mrz, and the size of one of these bands was larger than the predicted size of 27.9 kDa. A previous study has shown that the amino acid sequence of EtSAG10 has a high frequency of change between amino acid residues 140 and 160, which may change the spatial conformation and hydrophilicity of the protein (Qiu et al. 2017). This change in protein size may also be due to post-translational modifications, including acylation, alkylation, and methylation. A study on T. gondii found that rhoptry protein 2 and microneme protein 2 had two bands on Western blots, corresponding to a precursor form and an active form (Entzeroth et al. 1998).
Indirect immunofluorescence using an antibody against rEtSAG10 showed that the protein was mainly localized to the parasite surface. Furthermore, EtSAG10 expression was higher in Mrz than Spz, and this result was consistent with the Western blot results. EtSAG10 expression was increased as parasite development progressed in DF-1 cells. Parasite surface proteins are particularly interesting because of their potential role in resisting the external environment and adapting to new environments. For instance, the trypomastigote small surface antigen (TSSA) plays a critical role in the infectivity and differentiation of T. cruzi trypomastigotes and the phenotypic variability of parasite strains (Camara et al. 2017). Moreover, the expression of a parasite surface antigen may cause a protective immune response in the host (Tabares et al. 2004; Chow et al. 2011; Petitdidier et al. 2016). Major changes in gene expression of surface antigen 10 in MRR and DZR may be involved in pathogenesis and immune evasion. The immune evasion mechanism of parasites involves (1) constantly changing the antigenicity of the parasite, (2) reducing the host immune response, and (3) avoiding or resisting the site of immune attack (Saito et al. 2017; Yam and Preiser 2017; Belachew 2018). In addition, EtSAG10 was detected in the PVM. The parasitophorous vacuole is crucial to protect the parasite against the host cell environment, and the PVM is a major route of communication between the intracellular parasite and the host cell by allowing the exchange of metabolites between them (Entzeroth et al. 1998; Beyer et al. 2002). Therefore, we hypothesized that EtSAG10 might escape the host immune response to protect the parasite against the intracellular environment.
EtSAG10 might also be involved in host cell invasion because invasion inhibition assays indicated that pretreatment with the anti-rEtSAG10 polyclonal antibody reduced the capacity of sporozoites to invade DF-1 cells in vitro. In a previous study, TSSA served as an anchor for invading parasites (Canepa et al. 2012). EtSAG10 is one of the SAG family members. Proteins from all SAG subfamilies contain signal peptides and GPI anchors (Reid et al. 2014). These GPI-anchored proteins are present at several developmental stages in T. gondii, Plasmodium, Cryptosporidium, and Neospora, and are essential for the adhesion of parasites to host cells (Lekutis et al. 2001; Leal-Sena et al. 2018). The present results supported these findings. However, the changes in surface antigens upon drug-stimulated coccidial invasion of host cells need to be better explored.
Conclusions
In this study, a full-length EtSAG10 cDNA from E. tenella was cloned, expressed, and characterized. The mRNA level was higher in sporulated oocysts than at other developmental stages. The present results suggested that EtSAG10 may be important in pathogenesis and immune evasion and may play a fundamental role in the invasion of host cells by sporozoites in vitro. Importantly, the difference between drug-resistant and sensitive strains may be related to E. tenella drug resistance to maduramicin and diclazuril, and this differential expression and mutations may provide a basis for seeking a marker of coccidial resistance. However, the function of this molecule needs to be further explored.
References
Antony HA, Pathak V, Parija SC, Ghosh K, Bhattacherjee A (2016) Transcriptomic analysis of chloroquine-sensitive and chloroquine-resistant strains of Plasmodium falciparum: toward malaria diagnostics and therapeutics for global health. Omics 20(7):424–432. https://doi.org/10.1089/omi.2016.0058
Aosai F, Mun HS, Norose K, Chen M, Hata H, Kobayashi M, Kiuchi M, Stauss HJ, Yano A (1999) Protective immunity induced by vaccination with SAG1 gene-transfected cells against Toxoplasma gondii-infection in mice. Microbiol Immunol 43(1):87–91
Belachew EB (2018) Immune response and evasion mechanisms of Plasmodium falciparum parasites. J Immunol Res 2018:6529681. https://doi.org/10.1155/2018/6529681
Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav-Tal Y (2010) The life of an mRNA in space and time. J Cell Sci 123(Pt 10):1761–1774. https://doi.org/10.1242/jcs.062638
Beyer TV, Svezhova NV, Radchenko AI, Sidorenko NV (2002) Parasitophorous vacuole: morphofunctional diversity in different coccidian genera (a short insight into the problem). Cell Biol Int 26(10):861–871
Camara MLM, Canepa GE, Lantos AB, Balouz V, Yu H, Chen X et al (2017) The trypomastigote small surface antigen (TSSA) regulates Trypanosoma cruzi infectivity and differentiation. PLoS Negl Trop Dis 11(8):e0005856. https://doi.org/10.1371/journal.pntd.0005856
Canepa GE, Degese MS, Budu A, Garcia CR, Buscaglia CA (2012) Involvement of TSSA (trypomastigote small surface antigen) in Trypanosoma cruzi invasion of mammalian cells. Biochem J 444(2):211–218. https://doi.org/10.1042/bj20120074
Cardwell MM, Martinez JJ (2009) The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun 77(12):5272–5280. https://doi.org/10.1128/iai.00201-09
Chapman HD (1997) Biochemical, genetic and applied aspects of drug resistance in Eimeria parasites of the fowl. Avian Pathol 26(2):221–244. https://doi.org/10.1080/03079459708419208
Chen T, Zhang W, Wang J, Dong H, Wang M (2008) Eimeria tenella: analysis of differentially expressed genes in the monensin- and maduramicin-resistant lines using cDNA array. Exp Parasitol 119(2):264–271. https://doi.org/10.1016/j.exppara.2008.02.010
Chow YP, Wan KL, Blake DP, Tomley F, Nathan S (2011) Immunogenic Eimeria tenella glycosylphosphatidylinositol-anchored surface antigens (SAGs) induce inflammatory responses in avian macrophages. PLoS One 6(9):e25233. https://doi.org/10.1371/journal.pone.0025233
Doliwa C, Xia D, Escotte-Binet S, Newsham EL, Sanya JS, Aubert D et al (2013) Identification of differentially expressed proteins in sulfadiazine resistant and sensitive strains of Toxoplasma gondii using difference-gel electrophoresis (DIGE). Int J Parasitol Drugs Drug Resist 3:35–44. https://doi.org/10.1016/j.ijpddr.2012.12.002
Entzeroth R, Mattig FR, Werner-Meier R (1998) Structure and function of the parasitophorous vacuole in Eimeria species. Int J Parasitol 28(7):1015–1018
Gazzinelli RT, Denkers EY (2006) Protozoan encounters with toll-like receptor signalling pathways: implications for host parasitism. Nat Rev Immunol 6(12):895–906. https://doi.org/10.1038/nri1978
Geysen J, Ausma J, vanden Bossche H (1991) Simultaneous purification of merozoites and schizonts of Eimeria tenella (Apicomplexa) by Percoll flotation and assessment of cell viability with a double fluorescent dye assay. J Parasitol 77(6):989–993
Gould EN, Corbeil LB, Kania SA, Tolbert MK (2017) Evaluation of surface antigen TF1.17 in feline Tritrichomonas foetus isolates. Vet Parasitol 244:144–153. https://doi.org/10.1016/j.vetpar.2017.08.001
Gregson A, Plowe CV (2005) Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 57(1):117–145. https://doi.org/10.1124/pr.57.1.4
Han HY, Zhao QP, Chen ZG, Huang B (2004) Induction of diclazuril resistant strains and madumycin resistant strains of Eimeria tenella in the laboratory. Chin J Vet Sci 24(2):138–140
Han HY, Lin JJ, Zhao QP, Dong H, Jiang LL, Xu MQ, Zhu SH, Huang B (2010) Identification of differentially expressed genes in early stages of Eimeria tenella by suppression subtractive hybridization and cDNA microarray. J Parasitol 96(1):95–102. https://doi.org/10.1645/GE-2221.1
Han H, Kong C, Dong H, Zhu S, Zhao Q, Zhai Q, Liang S, Li S, Yang S, Huang B (2015) Molecular characterization and functional analysis of subunit 7 of eukaryotic initiation factor 3 from Eimeria tenella. Exp Parasitol 154:118–126. https://doi.org/10.1016/j.exppara.2015.04.002
Huang B, Zhao QP, Wu XZ, Shi TW, Chen ZG (1993) Study on the identification and pathogenicity of the pure species of Eimeria tenella Shanghai. Shanghai J Anim Husb and Vet Med 5:18–20
Jahn D, Matros A, Bakulina AY, Tiedemann J, Schubert U, Giersberg M, Haehnel S, Zoufal K, Mock HP, Kipriyanov SM (2009) Model structure of the immunodominant surface antigen of Eimeria tenella identified as a target for sporozoite-neutralizing monoclonal antibody. Parasitol Res 105(3):655–668. https://doi.org/10.1007/s00436-009-1437-6
Jia PL, Zhao ZM (2017) Impacts of somatic mutations on gene expression: an association perspective. Brief Bioinform 18(3):413–425. https://doi.org/10.1093/bib/bbw037
Jiang LL, Huang B, Han HY, Zhao QP, Dong H, Chen ZG (2005) Comparison of the proteome of the sporutated oocysts of Eimeria tenella diclazuril sensitive strain with diclazuril resistant strain. Chin J Biotech 21(3):435–439
Jiang L, Lin J, Han H, Zhao Q, Dong H, Zhu S, Huang B (2012) Identification and partial characterization of a serine protease inhibitor (serpin) of Eimeria tenella. Parasitol Res 110(2):865–874. https://doi.org/10.1007/s00436-011-2568-0
Kumar D, Singh R, Bhandari V, Kulshrestha A, Negi NS, Salotra P (2012) Biomarkers of antimony resistance: need for expression analysis of multiple genes to distinguish resistance phenotype in clinical isolates of Leishmania donovani. Parasitol Res 111(1):223–230. https://doi.org/10.1007/s00436-012-2823-z
Leal-Sena JA, Dos Santos JL, Dos Santos TAR, de Andrade EM, de Oliveira Mendes TA, Santana JO et al (2018) Toxoplasma gondii antigen SAG2A differentially modulates IL-1beta expression in resistant and susceptible murine peritoneal cells. Appl Microbiol Biotechnol 102(5):2235–2249. https://doi.org/10.1007/s00253-018-8759-1
Lekutis C, Ferguson DJ, Grigg ME, Camps M, Boothroyd JC (2001) Surface antigens of Toxoplasma gondii: variations on a theme. Int J Parasitol 31(12):1285–1292
Leng L, Luo M, Gao J, Shen LJ (2014) Study and application of surface antigen in tachyzoites of Toxoplasma gondii. Chin J Schi Contl 26(6):687–689
Liu Z, Xi D, Kang M, Guo X, Xu B (2012) Molecular cloning and characterization of Hsp27.6: the first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperones 17(5):539–551. https://doi.org/10.1007/s12192-012-0330-x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Med Sci 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Mbengue A, Bhattacharjee S, Pandharkar T, Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio JJ, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K (2015) A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 520(7549):683–687. https://doi.org/10.1038/nature14412
Mollinedo F, Martin-Martin B, Gajate C, Lazo PA (1998) Physiological activation of human neutrophils down-regulates CD53 cell surface antigen. J Leukoc Biol 63(6):699–706
Montero E, Rodriguez M, Gonzalez LM, Lobo CA (2008) Babesia divergens: identification and characterization of BdHSP-20, a small heat shock protein. Exp Parasitol 119(2):238–245. https://doi.org/10.1016/j.exppara.2008.01.020
Peek HW, Landman WJ (2003) Resistance to anticoccidial drugs of Dutch avian Eimeria spp. field isolates originating from 1996, 1999 and 2001. Avian Pathol 32(4):391–401. https://doi.org/10.1080/0307945031000121149
Perez-Morales D, Ostoa-Saloma P, Espinoza B (2009) Trypanosoma cruzi SHSP16: characterization of an alpha-crystallin small heat shock protein. Exp Parasitol 123(2):182–189. https://doi.org/10.1016/j.exppara.2009.06.019
Peroval M, Pery P, Labbe M (2006) The heat shock protein 90 of Eimeria tenella is essential for invasion of host cell and schizont growth. Int J Parasitol 36(10–11):1205–1215. https://doi.org/10.1016/j.ijpara.2006.04.006
Petitdidier E, Pagniez J, Papierok G, Vincendeau P, Lemesre JL, Bras-Goncalves R (2016) Recombinant forms of Leishmania amazonensis excreted/secreted promastigote surface antigen (PSA) induce protective immune responses in dogs. PLoS Negl Trop Dis 10(5):e0004614. https://doi.org/10.1371/journal.pntd.0004614
Qiu BF, Jing J, Dong RL, Song HY, Liu C, Zhu SX et al (2017) Cloning and analysis of SAG10 gene of Eimeria tenella Jiangsu strain. Jiangsu Agric Sci 2017(03):24–28
Reid AJ, Blake DP, Ansari HR, Billington K, Browne HP, Bryant J, Dunn M, Hung SS, Kawahara F, Miranda-Saavedra D, Malas TB, Mourier T, Naghra H, Nair M, Otto TD, Rawlings ND, Rivailler P, Sanchez-Flores A, Sanders M, Subramaniam C, Tay YL, Woo Y, Wu X, Barrell B, Dear PH, Doerig C, Gruber A, Ivens AC, Parkinson J, Rajandream MA, Shirley MW, Wan KL, Berriman M, Tomley FM, Pain A (2014) Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res 24(10):1676–1685. https://doi.org/10.1101/gr.168955.113
Saito F, Hirayasu K, Satoh T, Wang CW, Lusingu J, Arimori T, Shida K, Palacpac NMQ, Itagaki S, Iwanaga S, Takashima E, Tsuboi T, Kohyama M, Suenaga T, Colonna M, Takagi J, Lavstsen T, Horii T, Arase H (2017) Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors. Nature. 552(7683):101–105. https://doi.org/10.1038/nature24994
Shirley MW (1995) Eimeria species and strains of chickens. In: Coudert P, Eckert J, Braun R, Shirley MW (eds) Biotechnology – guidelines on techniques in coccidiosis research. The European Commission DGXII, Luxembourg City, pp 9–10
Shirley MW, Smith AL, Tomley FM (2005) The biology of avian Eimeria with an emphasis on their control by vaccination. Adv Parasitol 60:285–330. https://doi.org/10.1016/s0065-308x(05)60005-x
Sun M, Lu MX, Tang XT, Du YZ (2014a) Characterization and expression of genes encoding three small heat shock proteins in Sesamia inferens (Lepidoptera: Noctuidae). Int J Mol Sci 15(12):23196–23211. https://doi.org/10.3390/ijms151223196
Sun H, Wang L, Wang T, Zhang J, Liu Q, Chen P, Chen Z, Wang F, Li H, Xiao Y, Zhao X (2014b) Display of Eimeria tenella EtMic2 protein on the surface of Saccharomyces cerevisiae as a potential oral vaccine against chicken coccidiosis. Vaccine. 32(16):1869–1876. https://doi.org/10.1016/j.vaccine.2014.01.068
Tabares E, Ferguson D, Clark J, Soon PE, Wan KL, Tomley F (2004) Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins. Mol Biochem Parasitol 135(1):123–132
Thabet A, Honscha W, Daugschies A, Bangoura B (2017) Quantitative proteomic studies in resistance mechanisms of Eimeria tenella against polyether ionophores. Parasitol Res 116(5):1553–1559. https://doi.org/10.1007/s00436-017-5432-z
Tomley F (1997) Techniques for isolation and characterization of apical organelles from Eimeria tenella sporozoites. Med Sci 13(2):171–176. https://doi.org/10.1006/meth.1997.0509
Wang Z, Huang B, Dong H, Zhao Q, Zhu S, Xia W, Xu S, Xie Y, Cui X, Tang M, Men Q, Yang Z, Li C, Zhu X, Han H (2016) Molecular characterization and functional analysis of a novel calcium-dependent protein kinase 4 from Eimeria tenella. PLoS One 11(12):e0168132. https://doi.org/10.1371/journal.pone.0168132
Williams RB (2002) Anticoccidial vaccines for broiler chickens: pathways to success. Avian Pathol 31(4):317–353. https://doi.org/10.1080/03079450220148988
Wilson DW, Goodman CD, Sleebs BE, Weiss GE, de Jong NW, Angrisano F et al (2015) Macrolides rapidly inhibit red blood cell invasion by the human malaria parasite, Plasmodium falciparum. BMC Biol 13:52. https://doi.org/10.1186/s12915-015-0162-0
Yam XY, Preiser PR (2017) Host immune evasion strategies of malaria blood stage parasite. Mol BioSyst 13(12):2498–2508. https://doi.org/10.1039/c7mb00502d
Zhai Q, Huang B, Dong H, Zhao Q, Zhu S, Liang S, Li S, Yang S, Han H (2016) Molecular characterization and immune protection of a new conserved hypothetical protein of Eimeria tenella. PLoS One 11(6):e0157678. https://doi.org/10.1371/journal.pone.0157678
Zhou BH, Wang HW, Wang XY, Zhang LF, Zhang KY, Xue FQ (2010) Eimeria tenella: effects of diclazuril treatment on microneme genes expression in second-generation merozoites and pathological changes of caeca in parasitized chickens. Exp Parasitol 125(3):264–270. https://doi.org/10.1016/j.exppara.2010.01.022
Acknowledgments
We would like to thank all organizations which funded this work and all the teachers who cooperated in technical assistance.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFD0500302), the National Natural Science Foundation of China (Grant No. 31572266, No. 31672551), the National Sharing Service Platform for Parasite Resources (No. TDRC-22), and the Shanghai Minhang District talent development special funds.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Xing-Quan Zhu
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Alignment of EtSAG10 DNA sequences of DS, DZR and MRR. (PNG 135 kb)
Fig. S2
Nucleotide sequence of the full-length cDNA and deduced amino acid sequence of EtSAG10. N-glycosylation site is shown in red. Casein kinase II phosphorylation site is indicated by the wavy underline. A putative N-myristoylation site is underlined. A protein kinase C phosphorylation site is shown in blue. The threonine-rich domain is in a rectangle. (PNG 198 kb)
Rights and permissions
About this article
Cite this article
Liu, G., Zhu, S., Zhao, Q. et al. Molecular characterization of surface antigen 10 of Eimeria tenella. Parasitol Res 118, 2989–2999 (2019). https://doi.org/10.1007/s00436-019-06437-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06437-0