Abstract
Mobile hosts like birds occupy a wide array of habitats in which they encounter various vector and parasite faunas. If the infection probability for vector-borne parasites varies among seasons and biomes, a migratory life can critically influence the infections of a host. The growing body of literature on avian blood parasites suggests that host migrations do not only influence prevalence of infection but can also evoke higher infection intensities and increased parasite diversity in migrant compared to resident host species. We investigated the prevalence, intensity and diversity of Plasmodium and Haemoproteus infections in three closely-related and sympatrically breeding sparrow species with different migration strategies ranging from residential house sparrow and partially migratory tree sparrow to the obligate migratory Spanish sparrow. With a prevalence of 49%, the migratory Spanish sparrows were significantly less frequently infected than the resident house sparrows (82%). The partially migratory tree sparrow showed an intermediate prevalence of 60%. The parasitaemias were similar in all three host species and indicated mostly chronic but also few acute infections. While we found Plasmodium parasites in all three sparrow species, only Spanish sparrows were infected with Haemoproteus parasites in our study. With nine clearly identified parasite lineages in our study and the highest number of lineages per infected individuals (i.e. relative diversity), Spanish sparrows harboured the most diverse parasite fauna. Our results suggest that migration strategies can affect Plasmodium and Haemoproteus infections of sparrows resulting in a lower parasite prevalence and higher parasite diversity in migratory hosts—at least during our host’s breeding period. A general scope for all annual cycle periods and across various bird taxa remains to be elucidated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Host migration strategy—ranging from fully resident to obligate migratory—and habitat use of hosts are considered prime factors in shaping vector-borne parasite infections (Altizer et al. 2011; Sehgal 2015). The diversity of current migration strategies, in turn, is considered to be the result of differential parasite pressures in various habitats across the globe (O’Connor et al. 2018). The net effect of host migration on parasite prevalence is ambiguous: migration can facilitate transmission through dense multi-species aggregations on hotspots resulting in higher prevalence year-round (Altizer et al. 2011). Alternatively, a migratory lifestyle can also hamper transmission and reduce prevalence when migratory hosts move away from parasite-rich areas for periods of the annual cycle (Loehle 1995), when infected migrants experience a higher mortality compared to non-infected conspecifics (Bradley and Altizer 2005) or when infected individuals migrate spatially or temporally separated from uninfected individuals (Johns and Shaw 2016). Endurance flights have also been associated with changes in physiology and immune function (Eikenaar and Hegemann 2016), which could ultimately affect the susceptibility of avian migrants.
The effect of habitat on vector-borne parasite infections is more straightforward: Ambient humidity and temperature jointly set the developmental conditions of parasites and vectors (Valkiūnas 2005; Pérez-Rodríguez et al. 2013). Insect vectors vary greatly in their developmental requirements and therefore differ in their distribution. For instance, if high humidity and temperature favour the larval development of a certain dipteran vector (Jarošík et al. 2011), it would occur in higher abundance and result in a higher probability of transmission of the associated parasites in humid and warm habitats (Loiseau et al. 2012). As migration increases the annual geographic range, migration amplifies habitat effects. Migratory hosts encounter a greater diversity of habitats and vectors within the annual cycle and, depending on the adaptability of the host-parasite systems, can harbour a more diverse parasite fauna compared to resident species (Figuerola and Green 2000; Koprivnikar and Leung 2015).
Here, we investigate the influence of host migration strategy on the prevalence, intensity and diversity of infections with two genera of haemosporidian parasites in Southeastern European sparrows. We used comparable sample sizes from a single area during the breeding season of three closely related species, the house sparrow Passer domesticus (Linnaeus, 1758), the tree sparrow P. montanus (Linnaeus, 1758) and the Spanish sparrow P. hispaniolensis (Temminck, 1820), in order to minimise confounding effects of sampling effort, breeding habitat, season and host phylogeny. The house sparrow is a globally distributed rural and urban breeder with a resident lifestyle throughout Europe (Cramp 1994). Haemosporidian parasites in the house sparrow are well studied, with global prevalence of around 40%—ranging from 0% in island populations and in the north of Europe to over 80% in the south of continental Europe (Marzal et al. 2011). In the public reference database about haemosporidian parasites of birds, one can find 12 genetic lineages of Plasmodium and Haemoproteus identified so far for house sparrows in Southeast Europe (MalAvi database accessed on the 08/06/2018; Bensch et al. 2009). Tree sparrows are generally partial short-distance migrants (Cramp 1994), i.e. when a fraction within a local population is resident whereas another fraction migrates short distances. Prevalences range from 4% in Bulgaria (Shurulinkov 2005) to 37% in Spain (Ventim et al. 2012b) and, so far, four lineages of Plasmodium and Haemoproteus have been identified in Southeastern European tree sparrows. Finally, the Spanish sparrow shows a more complex migration strategy with fully resident populations in Western Europe and obligate migratory eastern populations (Summers-Smith 1988). Prevalence varied between 33% in Bulgaria (Dimitrov et al. 2010) and 80% in a mixed colony with Indian house sparrows (Passer d. indicus) in Kazakhstan (Valkiūnas 2005) and seven lineages have been recorded in Spanish sparrows in Southeast Europe. Data on intensity of haemosporidian infections (i.e. the percentage of infected erythrocytes, henceforth referred to as ‘parasitaemia’) of sparrows are generally scarce, with few exceptions for house and tree sparrows (Ventim et al. 2012a; Bichet et al. 2014).
If migration strategy affects blood parasite infections, we expect little differences between the resident house sparrows and the partially migratory tree sparrow but pronounced differences between them and the obligate migratory Spanish sparrow (Fig. 1). In particular, we hypothesise that (i) prevalence of haemosporidian infections in Spanish sparrows differs from those in house and tree sparrows: prevalence would be higher if migration increases susceptibility to blood parasite infections, or prevalence would be lower if migration hampers parasite transmission. Furthermore, we expect that (ii) parasitaemia would be higher in Spanish sparrows than in the other two species, if parasite intensities resulting from flight-induced relapses (Gylfe et al. 2000) or infections transmitted during migration remain elevated until the breeding season. Finally, we expect (iii) parasite diversity to be higher in Spanish sparrows compared to house and tree sparrows, provided migrant species do not only encounter but also get infected by more diverse parasites on their journeys.
Hypothesised influence of host migration strategy on parasite prevalence, parasitaemia and diversity—relative between three sympatrically breeding sparrow species (PasDom = house sparrow, PasMon = tree sparrow, PasHis = Spanish sparrow). We expect more distinctly differing infection parameters (double signs) in the obligate migratory than compared among the partially-migratory or resident sparrows (single signs)
Materials and methods
Study site and periods
We analysed haemosporidian infections in house sparrows, tree sparrows and Spanish sparrows breeding in the Danube river flood plain in Southeast Europe. We captured the sparrows using mist nets in the vicinity of the Biological Experimental Station ‘Kalimok’ and the village of Nova Cherna (Bulgaria; 44.00° N 26.45° E) in three consecutive periods in 2015, covering mid-breeding (July 1–18), late-breeding (July 25–August 10) and post-breeding (August 17–30).
Blood sampling
All birds were sexed (based on sex-specific plumage and cloacal protuberance), aged, weighed and ringed. To avoid misclassifications of juveniles and females between house and Spanish sparrows, we only sampled adults and preferably males, yielding sex ratios (males/females/unknown) of 39/7/1 for house sparrows, 18/5/27 for tree sparrows and 83/0/0 for Spanish sparrows (see Appendix A in ESM 1). In total, we sampled approximately 30 μl blood from 180 individuals (sample sizes per species and period were 16/16/15 for house sparrows, 16/26/8 for tree sparrows and 21/28/34 for Spanish sparrows) from the brachial vein with a heparinized capillary. We immediately prepared two to three thin blood smears per individual. The smears were dried fast using a fan, fixed on the same day for 5 min in 100% methanol as well as stained within 1 month for 1 h in 10% Giemsa’s solution.
Extraction and PCR
For 177 birds, there was residual blood in the capillary to be stored in SET buffer (0.015 M NaCl, 0.05 M Tris, 0.001 M EDTA, pH 8) at − 20 °C for genetic analysis. From these samples, we extracted DNA (DNeasy blood and tissue kit, Qiagen) and, to test for the presence of parasites of the genera Plasmodium and Haemoproteus, we performed a nested PCR developed by Hellgren et al. (2004). In brief, the product of a first PCR (3 min at 94 °C + [30s at 94 °C + 30s at 50 °C + 45 s at 72 °C] for 20 cycles + 10 min at 72 °C) using the primer pair HaemNF1 and HaemNR3 (detecting several haemosporidian genera) was used as a template in the second PCR (identical thermal profile but with 35 cycles instead of 20 cycles) with the primer pair HaemF and HaemR2 (detecting Haemoproteus and Plasmodium only). The samples were not molecularly tested for the presence of Leucocytozoon infections. Further, the PCR products were visualised in stained (GelRed, Biotium Inc.) 2% agarose gels. Parasite-negative samples and samples with unclear results of the first nested PCR (weak bands) were analysed twice to avoid false negative results. Finally, we yielded positive PCR results for 102 samples which were used to calculate species-specific prevalence and determine parasitaemia.
Prevalence estimation from the records on the MalAvi database
We also used MalAvi database entries to estimate an average prevalence for each host species, to compare these to the prevalences in our study. Therefore, we used entries of all lineages which have been detected in the three sparrow species in Southeast Europe (i.e. Albania, Bosnia, Bulgaria, Greece, Croatia, Macedonia, Moldavia, Montenegro, Romania, Serbia and Turkey (Eastern Thrace)). As the number of tested and infected individuals are entered per lineage and not all studies explicitly declare the number of mixed infections in the respective article, we conservatively estimated ranges of prevalence for each study, with a lower limit resulting from the assumption of the maximal possible number of co-infections and an upper limit resulting from the assumption of the absence of co-infections. Finally, we calculated a weighted mean (weighted by the sample size of the studies) among all lower and among all upper limits within each host species to get one average range for each sparrow species.
Microscopic measurement of parasitaemia
We microscopically determined parasitaemia (% infected erythrocytes) in PCR-positive birds by counting infected erythrocytes in 100 microscopic fields with 1000× magnification (Primo Star and AxioCam ERc5s, Carl Zeiss AG). The total number of inspected erythrocytes was extrapolated from five microscopy pictures (i.e. every 20th field, following Boone et al. 2010). No blood parasites of the genus Leucocytozoon was found in any of the blood smears we screened. In case we found no infected erythrocytes in the blood smears of PCR-positive samples which showed no signal in the chromatogram, we did not only exclude those from the analysis of lineage diversity, but also considered those false positive and corrected the infection status to uninfected (n = 3) for the calculation of prevalence. In case we found no infected erythrocytes (n = 5) or only extracellular stages (n = 4) in blood smears of samples with clear chromatograms, we set the parasitaemia to half the minimum detectable intensity (i.e. ½ infected erythrocytes/total inspected erythrocytes; see Hahn et al. 2018).
Sequencing and lineage determination
Parasite-positive PCR products were purified by enzymatic clean-up and hydrolysis of excess reagents (ExoSAP-IT kit, Thermofisher), sequenced with the forward primer (HaemF) using a sequencing kit (ABI BigDye® Terminator v3.1 Cycle, Applied Biosystems) for single-pass sequencing and the fluorescent-labelled fragments were analysed by electrophoresis in an DNA analyser (ABI 3730xl, Applied Biosystems) following the manufacturers protocols. Samples which yielded clear chromatograms with the forward primer were sequenced with the backward primer (HaemR2). The sequences were checked, edited (BioEdit software; Hall (1999)) and assigned to known lineages from the MalAvi database (Bensch et al. 2009) by a multi-sequence analysis (msa function and package; Bodenhofer et al. (2015)). From the 102 samples, we skipped lineage assignment for 10 which repeatedly showed no signal. From the 92 sequences with signal, we yielded 46 reads of full length (479 bp). Four of the full-length sequences did not match 100% with known lineages. These sequences are most similar (1–3 base-pairs difference) to two very closely related and widespread lineages, and the sequences of these potentially new lineages can be found in the MalAvi database and in GeneBank (accession no. MH909229 - MH909232). Therefore, 42 sequences entered the analysis of lineage diversity, while 50 sequences (18 from house sparrows, 14 from tree sparrows and 18 from Spanish sparrows) had to be excluded from the analysis of lineage diversity. These comprise of 43 sequences with 70–99% of the full length and seven samples with only 28–50% of the full length. But, in brief, 29 of 50 sequences showed 90–95% accordance with the most similar lineage from MalAvi database. None of these samples shared the unique Haemoproteus sequence between 400 and 430 base pairs and 25 of 29 were most similar to two ubiquitous lineages. The remaining 21 of 50 sequences contained larger gaps and therefore reach maximal similarities between 90 and 100%, often with the same degree of similarity with several lineages. Therefore, we also could not resolve mixed infections, even if sophisticated methods exist (Pérez-Tris and Bensch 2005). As our procedure occasionally reads mixed infections as single infections, the frequency of certain lineages might be underestimated, (Valkiūnas et al. 2006), but prevalence and parasitaemia are not affected by this imprecision.
For gaining insight on potential transmission areas of the parasites, we screened the MalAvi database for the lineages we detected in our three host species and made an overview on whether or not the lineage had been found in resident and/or migrant bird species in Southeast Europe, the rest of Europe and/or Africa, in order to identify presumed transmission areas.
Statistical analysis
We tested for differences in prevalence between hosts and sampling periods by fitting a Generalised Linear Mixed-Effects Model (function glmer of the R package lme4; Bates et al. (2015)) with infection status as dependent variable (family = binomial) and both species and periods as independent variables. To explicitly allow for species-specific intercept and slopes, we included species and periods in a random term (for modelling details, see Appendix B in ESM 2). Additionally, we calculated average prevalence per host species from the MalAvi database entries weighted for the varying sample sizes of the different studies.
Similar to the model for prevalence, we fitted a Linear Model to test for differences in parasitaemia (function lm of the R package stats; R Core Team (2016)) with the log of individual parasitaemia as dependent variable as well as species and period as independent variables (see Appendix B in ESM 2).
We tested for host-specific differences in lineage diversity by calculating relative diversity (= number of lineages/number of infected hosts) for both our study (with the actual number of infected hosts) and the lineages in the MalAvi database (with the estimated number of infected hosts i.e. the mean of the possible range of infected individuals). Relative diversity values depict the average number of lineages per infected host individual and account for differences in sample size per host species. Additionally, we calculated rarefaction (function rarefy), which corrects the number of found lineages accounting for rare lineages in our study and Chao dissimilarity indices (function vegdist (method = “chao”), both of the R package vegan; Oksanen et al. (2018)) which integrates the information about unique and shared lineages to compare parasite assemblages.
Data availability
All accompanying data on parasites are on the MalAvi database and along with the appendices on https://doi.org/10.17632/5gj8hkt4n2.1.
Results
Prevalence
In total, 82% of the house sparrows, 60% of the tree sparrows and 49% of the Spanish sparrows were infected with Plasmodium or Haemoproteus parasites (Fig. 2a). We detected Plasmodium infections in all three sparrow species but Haemoproteus infections only in Spanish sparrows. Thus, the prevalence was significantly higher in house sparrows compared to tree sparrows (GLME: slope = − 1.25[± 0.55]; z value = − 2.29; p value = 0.02) and Spanish sparrows (GLME: slope = − 1.54[± 0.48]; z value = − 3.22; p value = 0.001), but not significantly different between the latter two species (GLME: z = − 0.29, p = 0.46). Prevalence did not significantly change over the three periods (GLME: z values = 0.09–1.43, p values = 0.15–0.86). The prevalence deduced from the MalAvi database entries from Southeast Europe range from 35 to 60% in house sparrows, from 41 to 56% in tree sparrows and from 15 to 23% in Spanish sparrows.
Prevalence and parasitaemia of haemosporidian infections in the three sympatric sparrow species (PasDom = house sparrow, PasMon = tree sparrow, PasHis = Spanish sparrow): The prevalence (a) is significantly higher in house sparrows compared to tree sparrows and Spanish sparrows. Prevalence did not differ significantly between the three periods (grey bars: 1 = July 1–18, 2 = July 25–August 10, 3 = August 17–30, colour bars: average over the three periods). The parasitaemia (b) varied from 0.001 to 2% and differs neither between host species nor between periods (dots = individual parasitaemia, horizontal line = median, vertical line = 25–75% quartiles)
Parasitaemia
The infection intensities varied between 0.001 and 2% with a median of 0.01% (25th to 75th percentile 0.003–0.02%; n = 31) for house sparrows, 0.02% (0.004–0.03%; n = 27) for tree sparrows and 0.01% (0.003–0.03%; n = 41) for Spanish sparrows (Fig. 2b). Parasitaemia did not differ between host species (LM: t values = − 0.1–1.2, p values = 0.3–0.9) nor between the three periods (LM: t values = − 0.1–0.9, p values = 0.3–0.9).
Diversity
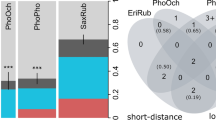
We assigned 42 sequences with 100% identity to 10 distinct cytochrome b lineages in the MalAvi database—seven lineages of Plasmodium and three of Haemoproteus (Fig. 3a). Two lineages (P-SGS1 and P-GRW11) are shared among all three sparrow species. House and Spanish sparrows additionally share one lineage (P-COLL1). Furthermore, house sparrows carry one (P-GRW06) unique lineages and Spanish sparrows six (H-PADOM05, H-PAMON01, H-PAHIS1, P-DELURB4, P-PADOM01 and P-PAGRI02). Many of the lineages have already been recorded for these three sparrow species in Southeast Europe (Fig. 3b). The Chao estimates which integrate total, shared and unique lineages indicate that the parasite assemblages of the house and tree sparrow are more similar to each other (Chao-dist PasDom-PasMon = 0.3) than to those of the Spanish sparrow (Chao-dist PasDom-PasHis = 0.63, Chao-dist PasMon-PasHis = 0.68). When dividing the number of lineages (4 for the house sparrow, 2 for the tree sparrow and 9 for the Spanish sparrow) by the number of infected individuals for each host species, we obtain relative diversities of 0.13 lineages per infected house sparrow, 0.07 lineages per infected tree sparrow and 0.22 for the Spanish sparrow. The relative diversities from the MalAvi database amount to 0.09 lineages per infected house sparrows, 0.07 lineages per infected tree sparrow and 0.30 lineages per infected Spanish sparrow. When correcting the total number of lineages per host for rare parasites, we obtain similar rarefied number of lineages (4 ± 0 for the house sparrow, 1.96 ± 0.2 for the tree sparrow and 6.4 ± 1 for the Spanish sparrow).
Plasmodium and Haemoproteus lineages found in house sparrows (PasDom), tree sparrows (PasMon) and Spanish sparrows (PasHis) from Southeast Europe. a Unique and shared cytochrome b lineages identified from the infected individuals in our study (sample sizes in brackets, northern Bulgaria, 1 year). b Lineage records from Southeast Europe (i.e. Albania, Bosnia, Bulgaria, Greece, Croatia, Macedonia, Moldavia, Montenegro, Romania, Serbia and Turkey (Eastern Thrace) archived in the MalAvi database (accessed on the 08/06/2018, sample size approximation in brackets). The prefix of the lineage abbreviation signifies the genus Plasmodium (P-) or Haemoproteus (H-). Note that a only includes the clearly assigned lineages of our study (sequences with 100% identity with a known lineage from the MalAvi database). The sizes of the coloured bubbles represent the absolute number of lineages found in each host species, but not relative diversity (= the number of lineages/the number of infected individuals)
Discussion
The migratory Spanish sparrows showed a significantly lower prevalence than the resident house sparrows. The intermediate prevalence of the partially migratory tree sparrow only differed from the house sparrow’s but not from the Spanish sparrow’s prevalence. In contrast, parasitaemia levels were similar in all three host species and Spanish sparrows are presumably infected with a higher diversity of haemosporidian parasites.
The absolute prevalence levels were generally higher in our study compared to those deduced from the MalAvi database, but the relative proportions are very similar. Parasite prevalence in Spanish sparrow was 40% lower than in house sparrows and 18% lower than in tree sparrows, which supports our hypothesis, that the prevalence of the house and tree sparrow will not differ to such a degree, compared to the prevalence of the Spanish sparrow (see Fig. 1). We acknowledge that our approach allows for a relative comparison but not for an absolute description of species-specific prevalence and its variability. As prevalence can vary between regions and years, spatial and temporal replicates of the sampling scheme are needed for such an endeavour. Unfortunately, we cannot disentangle if the lower prevalence in the Spanish sparrow is caused by increased mortality of infected compared to uninfected individuals on migration (known as “migratory culling”, Bradley and Altizer 2005) or if the migratory sparrows already started the journey with a lower pre-selective prevalence, which would mean that physiological or behavioural processes related to the host’s migratory lifestyle either changed the transmission probability of pathogens or the hosts’ susceptibility to parasites. Increasing migration distance along with an increasing time spent away from the breeding site has been shown to reduce the prevalence of locally transmitted pathogens in theoretical models of the transmission dynamics in animal populations (Hall et al. 2014).
Our data do not support the hypothesis of a higher parasitaemia in migratory hosts. Parasitaemia averages on similar level across the sparrows during the breeding period and seems to be unaffected by the hosts migration strategy. However, it cannot be excluded that endurance flights have induced relapses, but elevated parasitaemia did not persist until the breeding season. Yet, parasitaemia might in fact be mostly driven by seasonal changes in host physiology (Allander and Sundberg 1997), seasonal vector abundance and the influx of immunonaïve hosts after the reproductive season (Altizer et al. 2006).
Since our three host species are closely related and breed in close proximity, they probably share many vectors—at least during parts of the breeding period. Thus, the disparities in the sets of parasites might arise from transmission during the non-breeding season: Spanish sparrows have regularly been observed in Egypt and especially in the Nile delta during the non-breeding period (Summers-Smith 1988) and Spanish sparrows from our population likely spend the non-breeding period there. In these regions they encounter habitats with different climatic conditions and landscape elements (Pérez-Rodríguez et al. 2013), therefore parasites which cannot develop or be transmitted by vectors at the temperate breeding grounds (Žiegytė and Valkiūnas 2014).
The Chao estimates suggest that the parasite assemblages of the house and tree sparrow are more similar to each other compared to the assemblage of the Spanish sparrow. Furthermore, already before our study, migratory hosts were repeatedly found to harbour parasites of the genus Haemoproteus more often than resident hosts do (Clarabuch and Gonzalez-Solis 1997 and references therein). On the level of parasite assemblage and parasite genus diversity, this also supports our hypothesis, that the differences to the obligate migratory Spanish sparrow are expected to be the most pronounced compared to between the resident house sparrow and the partially migratory tree sparrow (see Fig. 1). As the vector taxa of Haemoproteus parasites are cosmopolitan, there is no trivial explanation for this pattern. If the higher parasite diversity in the Spanish sparrow would indeed originate from transmissions during the non-breeding period in Africa, we would expect lineages uniquely found in Spanish sparrows to be exclusively recorded in resident hosts in Africa but not in resident host species in Europe (Table 1). Yet, P-PAGRI02 was the only lineage in the MalAvi database that was exclusively detected in resident hosts in North Africa (Mata et al. 2015). The other lineages unique to Spanish sparrow in our study have also been detected in resident hosts in Southeast Europe and the rest of Europe (Table 1). So, except for P-PAGRI02, the MalAvi database offers little evidence that the surplus diversity of Spanish sparrows was transmitted in the non-breeding sites.
Alternative differing parasite assemblages could be explained if parasites require a certain period of time for local adaptation to a host-vector system. Then, recently established host populations would be expected to carry less locally transmitted parasites than a host species with long historical occupancy. In our study area, house and tree sparrows are traditional members of the avifauna and should thus carry more locally transmitted parasites compared to Spanish sparrows, which colonised the area just 50 years ago. But we cannot describe the complete lineage diversity of the three host species in this region, as a considerable number of sequences could not be assigned because they either did not reach 100% identity with a known lineage (n = 29) or shared maximal similarity with several known lineages (n = 7) or both (n = 14). To describe the complete haemosporidian diversity, one would require a larger sample per host species also balanced for sex and age classes and coverage of a longer period of the annual cycle over several years to level out the intra- and inter-annual variation.
However, we aimed for a temporal snapshot for the comparison of prevalence, parasitaemia and lineage diversity along a gradient of host migration strategies. The following peculiarities indicate that the unidentified lineages do not attenuate our findings of increased lineage diversity in the migratory Spanish sparrows: With 12 unidentifiable sequences stemming from Spanish sparrows, 6 from house sparrows and 5 from tree sparrows most of them origin from Spanish sparrows. All unidentified sequences of house and tree sparrows clustered with Plasmodium lineages. The majority of all unidentified sequences with maximal similarity with a single known lineage were most similar to the widespread lineages SGS1 (19 of 29) and GRW11 (8 of 29) and can thus already be found in the set of 100% matches (exception is one 94% match with P-ACAGR1 in a house sparrow and one 91% match with P-PADOM07 in a tree sparrow). Finally, we are confident that also the description of the four potentially new lineages would not change our result as three of the four sequences originate from Spanish sparrows and only one from a house sparrow. But to enable the description of the complete lineage diversity in the framework of future studies, the sequences are made available on a repository (see ‘Data accessibility’ section).
In conclusion, our findings of higher diversity, but lower prevalence in the migratory compared to the resident hosts support the idea that migration is a mixed blessing for the hosts of avian blood parasites. Host migration, on one side, incurs the cost of acquiring more diverse parasites, but can also come with benefits for the host. Theoretical work already indicated, that populations with a migratory life-style can profit from lower prevalence (Johns and Shaw 2016). However, the parasitism-related benefits of migration are rarely investigated empirically (but see Teitelbaum et al. 2018). Our study of three sympatrically breeding sparrow species covering a range of migration strategies from resident to obligate migratory provides the basic information for more extensive studies on the interaction of host migration strategy and blood parasites. In how far our findings can be generalised across bird taxa and the causal mechanisms behind such migration-related benefits still remain to be elucidated, for instance by investigating parasitism in populations with differing migratory propensity in a partially migratory species or in species pairs with varying migration strategies covering a broader range of taxa. Parasitism could even act as a selective pressure to increase or uphold migratory propensity, counter-balancing the expected selective pressure towards a reduction in migratory propensity in European bird communities caused by climate change (Schaefer et al. 2007), if the benefit of lower prevalence in migrants outrange the costs of hosting diverse parasites.
References
Allander K, Sundberg J (1997) Temporal variation and reliability of blood parasite levels in captive yellowhammer males Emberiza citrinella. J Avian Biol 28:325–330
Altizer SM, Bartel RA, Han BA (2011) Animal migration and infectious disease risk. Science 331:296–302. https://doi.org/10.1126/science.1194694
Altizer SM, Dobson AP, Hosseini P, Hudson P, Pascual M, Rohani P (2006) Seasonality and the dynamics of infectious diseases. Ecol Lett 9:467–484
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358. https://doi.org/10.1111/j.1755-0998.2009.02692.x
Bichet C, Sorci G, Robert A, Julliard R, Lendvai AZ, Chastel O, Garnier S, Loiseau C (2014) Epidemiology of Plasmodium relictum infection in the house sparrow. J Parasitol 100:59–65. https://doi.org/10.1645/12-24.1
Bodenhofer U, Bonatesta E, Horejš-Kainrath C, Hochreiter S (2015) msa: an R package for multiple sequence alignment. Bioinformatics 31:3997–3999. https://doi.org/10.1093/bioinformatics/btv494
Boone AT, Rodewald PG, DeGroote LW (2010) Neotropical winter habitat of the Magnolia warbler: effects on molt, energetic condition, migration timing, and hematozoan infection during spring migration. Condor 112:115–122. https://doi.org/10.1525/cond.2010.090098
Bradley CA, Altizer SM (2005) Parasites hinder monarch butterfly flight: implications for disease spread in migratory hosts. Ecol Lett 8:290–300. https://doi.org/10.1111/j.1461-0248.2005.00722.x
Clarabuch O, Gonzalez-Solis J (1997) Parasitism as a migration cost. Biol e Conserv della Fauna 102:113–117
Cramp S (1994) Handbook of the birds of Europe, the Middle East and North Africa: crows and finches. Oxford University Press, Oxford
Dimitrov D, Zehtindjiev P, Bensch S (2010) Genetic diversity of avian blood parasites in SE Europe: cytochrome b lineages of the genera Plasmodium and Haemoproteus (Haemosporida) from Bulgaria. Acta Parasitol 55:201–209. https://doi.org/10.2478/s11686-010-0029-z
Eikenaar C, Hegemann A (2016) Migratory common blackbirds have lower innate immune function during autumn migration than resident conspecifics. Biol Lett 12:20160078. https://doi.org/10.1098/rsbl.2016.0078
Figuerola J, Green AJ (2000) Haematozoan parasites and migratory behaviour in waterfowl. Evol Ecol 14:143–153. https://doi.org/10.1023/A:1011009419264
Gylfe Å, Bergström SS, Lundström J, Olsen B, Lundstróm J, Olsen B (2000) Reactivation of Borrelia infection in birds. Nature 403:724–725. https://doi.org/10.1038/35001663
Hahn S, Bauer S, Dimitrov D, Emmenegger T, Ivanova K, Zehtindjiev P, Buttemer WA (2018) Low intensity blood parasite infections do not reduce the aerobic performance of migratory birds. Proc R Soc B 285:20172307. https://doi.org/10.1098/rspb.2017.2307
Hall RJ, Altizer SM, Bartel RA (2014) Greater migratory propensity in hosts lowers pathogen transmission and impacts. J Anim Ecol 83:1068–1077. https://doi.org/10.1111/1365-2656.12204
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802. https://doi.org/10.1645/GE-184R1
Jarošík V, Honěk A, Magarey RD, Skuhrovec JJ, Honek A (2011) Developmental database for phenology models: related insect and mite species have similar thermal requirements. J Econ Entomol 104:1870–1876. https://doi.org/10.1603/EC11247
Johns S, Shaw AK (2016) Theoretical insight into three disease-related benefits of migration. Popul Ecol 58:213–221. https://doi.org/10.1007/s10144-015-0518-x
Koprivnikar J, Leung TLF (2015) Flying with diverse passengers: greater richness of parasitic nematodes in migratory birds. Oikos 124:399–405. https://doi.org/10.1111/oik.01799
Loehle C (1995) Social barriers to pathogen transmission in wild animal populations. Ecology 76:326–335. https://doi.org/10.2307/1941192
Loiseau C, Harrigan RJ, Robert A, Bowie RCK, Thomassen HA, Smith TB, Sehgal RNM, Francisco S, Angeles L, Angeles L, Africa S (2012) Host and habitat specialization of avian malaria in Africa. Mol Ecol 21:431–441. https://doi.org/10.1111/j.1365-294X.2011.05341.x
Marzal A, Ricklefs RE, Valkiūnas G, Albayrak T, Arriero E, Bonneaud C, Czirják GÁA, Ewen J, Hellgren O, Hořáková D, Iezhova TA, Jensen H, Križanauskienė A, Lima MR, de Lope F, Magnussen E, Martin LB, Møller AP, Palinauskas V, Pap PL, Pérez-Tris J, Sehgal RNM, Soler M, Szöllosi E, Westerdahl H, Zehtindjiev P, Bensch S, Zetindjiev P (2011) Diversity, loss, and gain of malaria parasites in a globally invasive bird. PLoS One 6:1–8. https://doi.org/10.1371/journal.pone.0021905
Mata VA, da Silva LPLP, Lopes RJ, Drovetski SV (2015) The Strait of Gibraltar poses an effective barrier to host-specialised but not to host-generalised lineages of avian Haemosporidia. Int J Parasitol 45:711–719. https://doi.org/10.1016/j.ijpara.2015.04.006
O’Connor EA, Cornwallis CK, Hasselquist D, Nilsson JÅ, Westerdahl H (2018) The evolution of immunity in relation to colonization and migration. Nat Ecol Evol 2:841–849. https://doi.org/10.1038/s41559-018-0509-3
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2018) vegan: community ecology package. R Package. Version 2:5–2
Pérez-Rodríguez A, Fernández-González S, de la Hera I, Pérez-Tris J (2013) Finding the appropriate variables to model the distribution of vector-borne parasites with different environmental preferences: climate is not enough. Glob Chang Biol 19:n/a-n/a. https://doi.org/10.1111/gcb.12226
Pérez-Tris J, Bensch S (2005) Diagnosing genetically diverse avian malarial infections using mixed-sequence analysis and TA-cloning. Parasitology 131:15–23. https://doi.org/10.1017/S003118200500733X
R Core Team (2016) R: A language and environment for statistical computing
Schaefer H-C, Jetz W, Böhning-Gaese K (2007) Impact of climate change on migratory birds: community reassembly versus adaptation. Glob Ecol Biogeogr 17:071106211200001–??? https://doi.org/10.1111/j.1466-8238.2007.00341.x
Sehgal RNM (2015) Manifold habitat effects on the prevalence and diversity of avian blood parasites. Int J Parasitol Parasites Wildl 4:421–430. https://doi.org/10.1016/j.ijppaw.2015.09.001
Shurulinkov P (2005) Occurrence of haematozoan parasites of genus Hepatozoon (Apicomplexa: Hepatozoidae) in wild birds in Bulgaria. Acta Zool Bulg 57:245–252
Summers-Smith JD (1988 The Sparrows : a study of the genus Passer. Calton, Staffordshire, England: T & AD Poyser
Teitelbaum CS, Huang S, Hall RJ, Altizer SM (2018) Migratory behavior predicts greater parasite diversity in ungulates. Proc R Soc B Biol Sci
Valkiūnas G (2005) Avian malaria parasites and other Haemosporidia. CRC Press, Boca Raton
Valkiūnas G, Bensch S, Iezhova TA, Krizanauskiené A, Hellgren O, Bolshakov CV (2006) Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. J Parasitol 92:418–422. https://doi.org/10.1645/GE-3547RN.1
Ventim R, Mendes L, Ramos JA, Cardoso H, Pérez-Tris J (2012a) Local haemoparasites in introduced wetland passerines. J Ornithol 153:1253–1259. https://doi.org/10.1007/s10336-012-0860-0
Ventim R, Tenreiro P, Grade N, Encarnação P, Araújo M, Mendes L, Pérez-Tris J, Ramos JA (2012b) Characterization of haemosporidian infections in warblers and sparrows at south-western European reed beds. J Ornithol 153:505–512. https://doi.org/10.1007/s10336-011-0767-11
Žiegytė R, Valkiūnas G (2014) Recent advances in vector studies of avian haemosporidian parasites. Ekologija 60:73–83. https://doi.org/10.6001/ekologija.v60i4.3042
Acknowledgments
We thank Bill Buttemer, Karina Ivanova, Martin Marinov and Strahil Peev for their support with field work as well as Zubera and Lyatif Ismail for their great hospitality. Finally, we would like to thank two anonymous reviewers for their constructive comments and inputs.
Funding
This study was funded by the Swiss National Science Foundation (SNSF) as a part of the project 31003A_160265. While TE, SB and SH were exclusively funded by SNSF, DD and PZ were co-funded from the Bulgarian National Science foundation under contract DN01/6 and JOM was funded by the Swiss Ornithological Institute. This study is report no. 63 of the Biological Experimental Station ‘Kalimok’.
Author information
Authors and Affiliations
Contributions
TE, SB and SH designed the study; PZ and DD arranged permissions and licences; TE, DD and SH organised and carried out the field work; JOM carried out the molecular lab work; TE and SH performed the screening by microscopy; TE implemented the data analysis and drafted the manuscript. All authors have revised the draft and agreed on the final version of the article.
Corresponding author
Ethics declarations
Ethical statement
All procedures were in accordance with the local animal ethics guidelines and were permitted by the Bulgarian Ministry of Environment and Waters (licence no. 574/27.03.2014).
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Bill Chobotar
Rights and permissions
About this article
Cite this article
Emmenegger, T., Bauer, S., Dimitrov, D. et al. Host migration strategy and blood parasite infections of three sparrow species sympatrically breeding in Southeast Europe. Parasitol Res 117, 3733–3741 (2018). https://doi.org/10.1007/s00436-018-6072-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6072-7