Abstract
Migration (seasonal round-trip movement across relatively large distances) is common within the animal kingdom. This behaviour often incurs extreme costs in terms of time, energy, and/or survival. Climate, food, predation, and breeding are typically suggested as factors favouring the evolution of migration. Although disease regulation has also been considered, few studies consider it as the primary selective pressure for migration. Our aim was to determine, theoretically, under what conditions migration could reduce the long-term disease prevalence within a population, assuming the only benefits of migration are infection-related. We created two mathematical models, one where the population migrates annually and one where the entire population remains on the breeding ground year-round. In each we simulated disease transmission (frequency-dependent and density-dependent) and quantified eventual disease prevalence. In the migration model we varied the time spent migrating, disease-related migration mortality, and the overall migration mortality. When we compared results from the two models, we found that migration generally lowered disease prevalence. We found a population was healthier if it: (1) spent more time migrating (assuming no disease transmission during migration), (2) had higher disease-induced migration mortality, and (3) had an overall higher mortality when migrating (compared to not migrating). These results provide support for two previously proposed mechanisms by which migration can reduce disease prevalence (migratory escape and migratory cull), and also demonstrate that non-selective mortality during migration is a third mechanism. Our findings indicate that migration may be evolutionarily advantageous even if the only migratory benefit is disease control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Migration is a behaviour that has evolved across a broad range of taxonomic groups (Alerstam et al. 2003; McGuire and Fraser 2014). Migration is characterised as an animal’s persistent movement between two or more habitats (Dingle 1996) and typically involves high energetic demands and extreme physiological changes (Lidicker and Caldwell 1982; Altizer et al. 2011). Migration can occur in numerous ways from long-range to-and-fro seasonal migration, as seen in the finetooth shark, Carcharhinus isodon (Castro 1983), to single one-way migration, as shown by the North American tumbleweed (Dingle 1996). In order to evolve such a complex and costly behaviour, migration must be highly beneficial. Many people have theorised about the exact nature of these benefits. The three main hypothesised benefits are increased food quality, finding mates or breeding sites, and avoiding the energetic costs associated with seasonally harsh climate or predation (Heape 1931; Aidley 1981; Avgar et al. 2014). Although reducing the energetic costs associated with parasites and pathogens is sometimes mentioned as a benefit of migration (e.g., Avgar et al. 2014), it is rarely studied as a primary factor driving migration (but see Poulin et al. 2012).
Many unfavourable diseases infect species that go through annual migration (Table 2 in Altizer et al. 2011), and it is clear that migration and infection could potentially interact in a number of ways. As individuals migrate, they can transfer pathogens to new areas (Reed et al. 2003). Infection by pathogens can also interfere with an individual’s ability to successfully migrate (van Gils et al. 2007). Furthermore, studies have shown that in some species migrants have a higher level of infection than residents (van Dijk et al. 2014) while in other species the reverse is true: migrants have a lower level of infection than residents (Folstad et al. 1991).
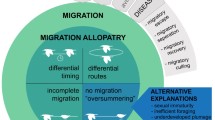
Three mechanisms have been proposed to explain how migration can reduce infection: migratory escape (Loehle 1995), migratory allopatry (Krkošek et al. 2007), and migratory cull (Bradley and Altizer 2005). Migratory escape occurs when individuals temporarily leave a highly infectious environment via migration. Pathogens that rely on hosts being present decline in number, resulting in lower virulence upon the individuals’ return to the environment. Similarly, migratory allopatry occurs when susceptible juveniles are physically separated from infectious adults (Krkošek et al. 2007). Migratory cull, on the other hand, occurs when infected individuals are not as capable of surviving migration, and thus reduce the prevalence of a disease within a population. Some studies have suggested that this process of natural selection against the infected leads to stronger immunity in migratory populations (Møller and Erritzøe 1998).
Although there is strong empirical support for each of these mechanisms, directly quantifying the disease control benefits of migration is difficult since migration can be driven by multiple covarying factors (e.g., disease and climate). However, such quantifications can be possible in theoretical studies of migration and disease. Hall et al. (2014) created a model to examine the interaction between disease transmission and seasonal migration, using life history assumptions typical of migratory Neotropical passerine species. Individuals in their theoretical population either remained at the breeding ground year-round or migrated to a wintering ground. They found that more extreme migration lowered disease prevalence (travelling longer distances and departing earlier from the infected breeding ground) and suggested that migration could therefore be adaptive for pathogen avoidance.
Here we develop a model for migration and disease that is similar to the approach in Hall et al. (2014), but with two key differences. First, we assume that the only benefits of migration are related to infection. In contrast, Hall et al. (2014) also assume seasonality in survival that differs between the breeding and non-breeding grounds. Second, we compare the infection prevalence in migratory and non-migratory populations, whereas Hall et al. (2014) compare the infection prevalence across different types of migratory populations. These two differences enable us to determine conservatively whether migration can minimise disease prevalence (maximise population health) if disease control is the only benefit to migration. We develop a pair of theoretical models and use them to compare disease prevalence with and without migration, considering both frequency-dependent and density-dependent disease transmission. We find that migration generally lowered disease prevalence and we demonstrate three mechanisms by which this occurs. Our findings suggest that disease control by itself could be a selective pressure for migration.
Methods
Here we model disease prevalence in a population of individuals that annually migrate between two areas: one where breeding occurs and a second, non-breeding, area. For simplicity we refer to the second as the ‘wintering grounds’, even though we do not assume seasonality in survival. We separate the year into four discrete time periods: breeding, migration, wintering, and return migration (Fig. 1). For comparison, we also consider a non-migratory population that spends the entire year on the breeding ground.
Schematic of the model’s annual cycle showing susceptible (S) and infected (I) individuals during the breeding, migration, wintering, and return migration time periods. Disease transmission (β) and recovery (γ) occur primarily on the breeding and wintering grounds. The mortality of susceptible and infected individuals are given by μ S and μ I (occurring continuously during the breeding and wintering seasons) and 1 − σ S and 1 − σ I (occurring once during each migration). The number of new offspring is given by b
Breeding and wintering grounds
We assume disease transmission occurs primarily in the breeding and wintering grounds. To model disease transmission between individuals, we employ a standard SI disease model, and we considered both frequency-dependent disease transmission
as well as density-dependent disease transmission
(for a full list of parameters and variable with their definitions and values see Table 1).
This splits the population into two groups: susceptible (S), healthy individuals, and infectious (I), individuals who have become infected by a disease. The rates at which individuals move between susceptible and infected classes depend both on how infectious the disease is (β′ and β) and how fast infected individuals recover (γ). We also assume that there is a constant disease-independent mortality rate, which is higher for infected individuals (μ I) than susceptible ones (μ S). Extra disease-related mortality occurs when there is the additional stress of migration (see below).
Migration
For simplicity, we assume that essentially no disease transmission occurs during migration. Each individual undertaking migration survives with probability σ, the likelihood of it reaching its destination. Since migration is typically quite costly, we assume that mortality during the migration period (length t M) is higher for a migrating individual than for one remaining on the breeding ground (\(1 - \sigma > e^{{ - \mu t_{\text{M}} }}\)).
To test the impact of migratory cull on disease prevalence, we varied the disease-related mortality cost (c) of having the disease. The survival of infected individuals (σ I) is given by
where σ S is the survival of healthy individuals. If there is no cost to the disease during migration, then c = 0 and healthy and infected individuals have equal survival probability during migration (\(\sigma_{\text{S}} = \sigma_{\text{I}}\)). However, if there is a cost (0 < c ≤ 1) then infected individuals have lower survival than healthy ones. Clearly this cull would initially reduce the number of infected individuals, but it is unclear if the disease would be restored to its previous level after migration ends and transmission resumes.
One year in the life
To create a basic annual structure, we used data from Setophaga coronata, the Yellow-Rumped Warbler, a well-studied migratory bird of the Americas. Note, however, that our model framework is not constrained to this species or even just to birds—it is general enough to be broadly applicable to migratory species across taxonomic groups. The Yellow-Rumped Warbler spends about 115 days on its breeding ground, 61 days migrating in the fall, 136 days on its wintering ground, and 53 days migrating in the spring (Burton 1992).
Each simulation was run as follows. The population is initialised with 10 % infected individuals (I 0 = 100, S 0 = 900). For the next t B days the population is on the breeding ground and experiences continuous disease transmission. At the end of this period the population migrates for t M days. We varied the relative disease-related mortality cost between susceptible and infectious individuals in order to quantify the effect of a migratory cull on the population.
Once the population arrives at the wintering grounds, disease transmission resumes. They remain here for t W days (t B + 2t M + t W = 365) and then spend t M days migrating back to their breeding ground. The population reproduces instantaneously at the beginning of the breeding time period. We assume reproduction is density-dependent, with the number of newborns given by
where r is the potential number of offspring an individual can have and K is the carrying capacity of the environment (the maximum number of individuals the environment can sustain). We assume that all newborns enter the susceptible category (there is no vertical transmission of the disease between parents and offspring).
For short-term dynamics, we report the disease prevalence (fraction of individuals that are infected) over time. For long-term dynamics, we focus on the disease prevalence immediately before reproduction at the start of the year. We ran each simulation for 100 years, until the disease prevalence at this point in the annual cycle did not change from year to year. We refer to this steady-state disease prevalence value as the ‘eventual disease prevalence’ for the population.
We fixed the ratio of transmission to recovery rates (β′/γ = 2 and β/γ = 2) but varied transmission rate for each frequency-dependent transmission (β′ = 0.01, 0.02, 0.03, and 0.05) and density-dependent transmission (β = 0.00001, 0.00002, 0.00003, and 0.00005), as well as disease-related mortality cost during migration (0 ≤ c ≤ 1). For comparison, we also ran an additional model where the population does not migrate. This model is simply the SI models (1) and (2) run throughout the entire year with births at the beginning of the year. We compared the eventual disease prevalence to determine how parameter values impacted the disease prevalence.
Results
Within year dynamics
Throughout the breeding period, the number of susceptible individuals decreased and the number of infected individuals increased (Fig. 2a–f). The rate at which this happened was affected by the value of β′, which represents the infectivity of the disease. When β′ was large (Fig. 2c, f), the disease spread quickly and the population rapidly reached a steady state. In contrast, in simulations with a smaller β′, disease spread slowly and the population typically did not reach the steady state before migration (Fig. 2a, d). If the steady state was not reached prior to migration, then the ‘migratory cull’ (increased mortality during migration) had the greatest impact, as the disease was still at relatively low levels in the population. This means that for a disease with a small β′, at the beginning of the migration there will be fewer infected individuals and thus the potential for the cull to ‘nip the disease in the bud’. Generally, the smaller the β′ value, the lower the disease prevalence at the end of the year. The results were qualitatively the same for density-dependent transmission (not shown).
Simulation of disease dynamics over time, showing the difference between one and multiple years as well as migratory versus non-migratory populations and different transmission rates, β′. The solid lines indicate the number of susceptible individuals (S), the dashed lines indicate the number of infected individuals (I), and grey bars indicate time periods spend migrating (no transmission). All simulations were run with disease-related migration mortality cost c = 0.3
Across year dynamics
As with the single year example, β′ changed the eventual (long-term) disease prevalence when the model was run over multiple years (Fig. 2g–l). A disease within a resident (non-migrating) population eventually had a 50 % prevalence (Fig. 2j–l) whereas the migrating population had much lower disease prevalence (Fig. 2g–i). For small β′, the disease eventually disappeared (Fig. 2g). We attributed this difference in disease prevalence to three factors that are present in the migration model but not in the resident model.
First, when there was a mortality cost to having the disease during migration (c > 0), disease prevalence was lower after the migratory cull in any given year. This resulted in a higher proportion of healthy individuals in the long-term. These results hold across multiple transmission rates which all showed a decrease in disease prevalence when the cost is increased, for both frequency-dependent (Fig. 3a) and density-dependent (Fig. 3b) transmission. The sharpest decline of infection was seen in the models with lower frequency-dependent transmission rates. The proportion of infected individuals was generally higher under density-dependent transmission than under frequency-dependent transmission, especially for low disease-related mortality.
Effect of disease mortality during migration. Eventual disease prevalence within the population as a function of disease-related migration mortality cost (c) for different transmission rates, for a frequency-dependent and b density-dependent transmission. As the disease becomes more costly, infected individuals are less likely to survive migration. Over multiple years this migratory cull increases the proportion of susceptibles within the population thus reducing eventual disease prevalence
Second, the migrating population in our model experienced a time of no transmission (since we assumed no transmission during migration) which overall reduced disease prevalence. We varied the time that migration takes (t M) and found that the more time spent migrating the lower the disease prevalence in the population (Fig. 4). This effect was much stronger for frequency-dependent transmission than for density-dependent transmission. In simulations with no migration, the proportion of infected individuals was approximately 0.5 when β′ was large (0.02, 0.03, and 0.05) and 0.47 when β′ was 0.01 (for frequency-dependent transmission) and the proportion of infected individuals was approximately 1.0 when β was large (0.00002, 0.00003, and 0.00005) and 0.96 when β was 0.00001 (for density-dependent transmission). Therefore, even for no time spent migrating (t M = 0), the disease prevalence was still higher in migration simulations than in the non-migrating ones.
Effect of non-transmission period. Eventual disease prevalence in the population as a function of time (in days) spent migrating (t M) for different transmission rates, for a frequency-dependent and b density-dependent transmission. The more time spent migrating the fewer infected individuals in the population. Here, there is no cost to having the disease during migration (c = 0). Even when migration takes no time there is still less infected individuals than in a non-migrating population
Third, any degree of mortality during migration (regardless of infection) led to a long-term reduction in disease prevalence. To separate out this third factor, we ran a set of simulations where there was no disease-related migration mortality cost (c = 0) and no time spent migrating (t M = 0). Here, we found that the higher the total migration mortality, the lower the proportion of infected individuals (Fig. 5). The change in infection prevalence with increasing mortality was much more drastic for density-dependent transmission (Fig. 5b) than for frequency-dependent transmission (Fig. 5a). The long-term disease prevalence was reduced even though the fraction of individuals dying during migration was the same for both susceptible and infected.
Effect of total migration mortality. Eventual disease prevalence after an instantaneous population decline (t M = 0) as a function of mortality probability during migration (1 − σ S, where c = 0 so σ = σ I = σ S) for different transmission rates, for a frequency-dependent and b density-dependent transmission. Here in the extreme case (1 − σ = 0), the eventual disease prevalence is the same for a migrating and non-migrating population
Discussion
Consistent with prior theoretical and empirical work, here we show that migration can benefit host populations by lowering disease prevalence. The novelty of our results comes from the fact that, in contrast to previous theoretical work, we have constructed a model where the only benefits of migration are lowered disease prevalence (e.g., no increased survival). We demonstrate that migration could indeed lower disease prevalence in a population, by acting through three mechanisms.
First, if infected individuals have lower survival during migration than healthy individuals do, disease prevalence is lower. Although a higher mortality of diseased individuals intuitively lowers disease prevalence in the short term, the long-term outcomes are less obvious. We found that when the infection rate is high, the disease prevalence quickly increases again after migration. In contrast, we show that when the infection rate is low, disease prevalence does not increase to the same level that was present prior to migration and, in the extreme, eventually dies out. As a result, migratory populations have lower disease prevalence than resident populations (an example of migratory cull; Bradley and Altizer 2005). The validity of a disease cull is supported by empirical evidence that migratory birds infected with low-pathogenic avian influenza (LPAI) can have deteriorated body condition (Latorre-Margalef et al. 2009), or travel shorter distances during migration (van Gils et al. 2007) compared to uninfected migrants. When the immune response is triggered it leads to a decrease in body condition (Nebel et al. 2012) thus reducing an individual’s chance of surviving migration. The same physiological impact can also be seen in individuals carrying parasites (Wagner et al. 2003; Bradley and Altizer 2005). As long as the parasite’s life cycle does not change the transmission dynamics, our model also applies to microparasite infection.
Second, when there is more time spent migrating, there is less time at the wintering and breeding grounds, where disease transmission occurs, which results in lower disease prevalence. This outcome is based upon our assumption of no disease transmission during migration, and this is effectively the ‘migratory escape’ mechanism proposed by Loehle (1995) and for which Hall et al. (2014) also found theoretical support. The assumption of no (or at the very least, lower) disease transmission would be more appropriate for species that form particularly dense aggregations on either the breeding or non-breeding grounds (e.g., elk, Cervus elaphus; Cross et al. 2010, and many reef fish; Domeier and Colin 1997). However, species like red knots (Calidris canutus) that migrate in groups or aggregate at stopover sites may actually experience increased transmission during migration (Buehler and Piersma 2008). The Hall et al. (2014) model assumes that transmission does not occur in the wintering ground (in contrast to our assumption of transmission in both breeding and wintering grounds), which may be the case in some species. Again, as the model developed by Hall et al. (2014) assumes that migration is beneficial even with no pathogen present, it is used to answer a different question (for a migrating species, what amount of time spent migrating and distance travelled is most beneficial?) than the one we answer here (when is migration more beneficial than not migrating?).
Last, the overall population decline caused by migration mortality (irrespective of an individual’s infection status) causes a reduction in disease prevalence. To measure this, we considered the case where migration is instantaneous and all individuals have the same chance of surviving migration. Here, higher mortality during migration reduces the mean lifespan of both susceptible and infected individuals. A reduced lifespan of infected individuals creates a lower probability of infecting susceptible individuals, and therefore reduces overall disease prevalence. This type of instantaneous cull could occur in situations other than migration, such as seasonal bushfires, storms, frosts or heatwaves that kill part of a population. In each case, we expect the cull to result in lower disease prevalence. In fact, this sort of non-selective culling has been used as a strategy for actively managing diseases such as bovine tuberculosis and chronic wasting disease in wildlife populations (Schmitt et al. 2002; Potapov et al. 2012; Manjerovic et al. 2014). Future empirical studies should be conducted to test whether natural events lead to lower disease prevalence as well. For example, studies could examine the effect of a single population drop and how large it could be before it becomes detrimental to the population.
We ran our model using both frequency-dependent and density-dependent transmission. The transmission of most diseases is likely somewhere in the middle so these should be seen as two ends of a spectrum rather than two completely distinct scenarios (Antonovics et al. 1995). For both transmission cases, migration reduced disease prevalence under all three of the mechanisms described above. However, the relative effect of each mechanism differed by transmission type. Increased time without disease transmission (time spent migrating in our model) had a bigger impact for frequency-dependent transmission. In contrast, increased mortality during migration (both disease-related mortality, and overall mortality) had a bigger impact for density-dependent transmission. Full eradication of the disease from the population was most feasibly under density-dependent transmission with increased overall mortality during migration, and under frequency-dependent transmission with increased disease-related mortality during migration. This matches standard understanding in disease modelling that non-selective reduction of population size is a feasible strategy for eradicating disease under density-dependent transmission, but not under frequency-dependent transmission (McCallum et al. 2001).
Although our model does show improved population health due to migration it does not track individuals and thus cannot determine if disease control could cause the evolution of migration. Future models should be made to test the evolutionary weight of disease control and determine if it could instigate migration. Empirical studies have shown that in some species, individuals decide to migrate or not based on body condition, with evidence for both better (Brodersen et al. 2008) and worse (Olsson et al. 2006) condition causing individuals to migrate. Perhaps if given the chances to remain behind, infected individuals would not migrate.
Since our model does not include migration distances explicitly, it could also be used to model any behaviour that involves a period of time when there is no disease transmission. For example, in the case of hibernation, disease transmission likely occurs throughout the year except during the time of hibernation (for species that hibernate solitarily). This system could also involve a cull for infected individuals as it could be hypothesised that the cost of the immune response would use energy needed to survive over the time of hibernation.
In addition to there being disease-related benefits of migration (explored here), there can also be disease-related costs of migration (not explored here). For example, in some species, migrating can actually increase exposure to pathogens (Figuerola and Green 2000; Waldenström et al. 2002; Morgan et al. 2007). In these cases, for migration to be favoured there must be some non-disease benefit to migration. Future modelling work could determine when the benefits of migration outweigh the costs, in the case where migration increases disease exposure.
Our model showed a decline in disease prevalence caused by three different factors: migratory cull of infected individuals, escaping transmission time via migratory trip, and a general population cull. The largest reduction in disease prevalence was caused by migratory cull. This is one of the first models to consider disease control as the only benefit to migration. Since we found such a large population benefit, we argue that disease control should be considered alongside climate, food, predation, and reproduction as potential benefits to migration.
References
Aidley DJ (1981) Questions about migration. In: Aidley DJ (ed) Animal migration. Cambridge University Press, Cambridge, pp 1–7
Alerstam T, Hedenström A, Akesson S (2003) Long-distance migration: evolution and determinants. Oikos 103:247–260
Altizer S, Bartel R, Han BA (2011) Animal migration and infectious disease risk. Science 331:296–302
Antonovics J, Iwasa Y, Hassell M (1995) A generalized model of parasitoid, venereal, and vector-based transmission processes. Am Nat 145:661–675
Avgar T, Street G, Fryxell JM (2014) On the adaptive benefits of mammal migration. Can J Zool 92:481–490
Bradley CA, Altizer S (2005) Parasites hinder monarch butterfly flight: implications for disease spread in migratory hosts. Ecol Lett 8:290–300
Brodersen J, Nilsson PA, Hansson L-A, Skov C, Brönmark C (2008) Condition-dependent individual decision-making determines cyprinid partial migration. Ecology 89:1195–1200
Buehler DM, Piersma T (2008) Travelling on a budget: predictions and ecological evidence for bottlenecks in the annual cycle of long-distance migrants. Philos Trans R Soc B-Biol Sci 363:247–266
Burton R (1992) Bird migration. Aurum press, London
Castro JI (1983) The sharks of North American waters. University Press, College Station
Cross PC, Cole EK, Dobson AP, Edwards WH, Hamlin KL, Luikart G, Middleton AD, Scurlock BM, White PJ (2010) Probable causes of increasing brucellosis in free-ranging elk of the Greater Yellowstone Ecosystem. Ecol Appl 20:278–288
Dingle H (1996) Migration: the biology of life on the move. Oxford University Press, Oxford
Domeier ML, Colin PL (1997) Tropical reef fish spawning aggregations: defined and reviewed. Bull Mar Sci 60:698–726
Figuerola J, Green AJ (2000) Haematozoan parasites and migratory behaviour in waterfowl. Evol Ecol 14:143–153
Folstad I, Nilssen AC, Halvorsen O, Andersen J (1991) Parasite avoidance: the cause of post-calving migrations in Rangifer? Can J Zool 69:2423–2429
Hall RJ, Altizer S, Bartel RA (2014) Greater migratory propensity in hosts lowers pathogen transmission and impacts. J Anim Ecol 83:1068–1077
Heape W (1931) Emigration, migration and nomadism. Heffer and Sons, Cambridge
Krkošek M, Gottesfeld A, Proctor B, Rolston D, Carr-Harris C, Lewis MA (2007) Effects of host migration, diversity and aquaculture on sea lice threats to Pacific salmon populations. Proc R Soc B-Biol Sci 274:3141–3149
Latorre-Margalef N, Gunnarsson G, Munster VJ, Fouchier RAM, Osterhaus ADME, Elmberg J, Olsen B, Wallensten A, Haemig PD, Fransson T, Brudin L, Waldenström J (2009) Effects of influenza A virus infection on migrating mallard ducks. Proc R Soc B-Biol Sci 276:1029–1036
Lidicker WZ, Caldwell RL (1982) Dispersal and migration. Hutchinson Ross Publishing Company, Stroudsburg
Loehle C (1995) Social barriers to pathogen transmission in wild animal populations. Ecology 76:326–335
Manjerovic MB, Green ML, Mateus-Pinilla N, Novakofski J (2014) The importance of localized culling in stabilizing chronic wasting disease prevalence in white-tailed deer populations. Prev Vet Med 113:139–145
McCallum H, Barlow N, Hone J (2001) How should pathogen transmission be modelled? Trends Ecol Evol 16:295–300
McGuire LP, Fraser EE (2014) Taxonomic diversity in the biology of migration. Can J Zool 92:463–465
Møller AP, Erritzøe J (1998) Host immune defence and migration in birds. Evol Ecol 12:945–953
Morgan ER, Medley GF, Torgerson PR, Shaikenov BS, Milner-Gulland EJ (2007) Parasite transmission in a migratory multiple host system. Ecol Model 200:511–520
Nebel S, Bauchinger U, Buehler DM, Langlois LA, Boyles M, Gerson AR, Price ER (2012) Constitutive immune function in European starlings, Sturnus vulgaris, is decreased immediately after an endurance flight in a wind tunnel. J Exp Biol 215:272–278
Olsson I, Greenberg LA, Bergman E, Wysujack K (2006) Environmentally induced migration: the importance of food. Ecol Lett 9:645–651
Potapov A, Merrill E, Lewis MA (2012) Wildlife disease elimination and density dependence. Proc R Soc B-Biol Sci 279:3139–3145
Poulin R, Closs GP, Lill AWT, Hicks AS, Herrmann KK, Kelly DW (2012) Migration as an escape from parasitism in New Zealand galaxiid fishes. Oecologia 169:955–963
Reed KD, Meece JK, Henkel JS, Shukla SK (2003) Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin Med Res 1:5–12
Schmitt SM, O’Brien DJ, Bruning-Fann CS, Fitzgerald SD (2002) Bovine tuberculosis in Michigan wildlife and livestock. Ann N Y Acad Sci 969:262–268
van Dijk JGB, Hoye BJ, Verhagen JH, Nolet BA, Fouchier RAM, Klaassen M (2014) Juveniles and migrants as drivers for seasonal epizootics of avian influenza virus. J Anim Ecol 83:266–275
van Gils JA, Munster VJ, Radersma R, Liefhebber D, Fouchier RAM, Klaassen M (2007) Hampered foraging and migratory performance in swans infected with low-pathogenic avian Influenza A virus. PLoS ONE 2:e184
Wagner GN, McKinley RS, Bjorn PA, Finstad B (2003) Physiological impact of sea lice on swimming performance of Atlantic salmon. J Fish Biol 62:1000–1009
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U (2002) Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol 11:1545–1554
Acknowledgments
We thank the ANU Theory Group for discussion of ideas, and S. Binning, S. Peacock, and two anonymous reviewers for helpful comments on previous versions of the manuscript. This material is based upon work supported by the National Science Foundation under Grant No. OISE-1159097 to AKS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Johns, S., Shaw, A.K. Theoretical insight into three disease-related benefits of migration. Popul Ecol 58, 213–221 (2016). https://doi.org/10.1007/s10144-015-0518-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-015-0518-x