Abstract
Acanthamoeba keratitis (AK) is a progressive corneal infection that demands rapid and sensitive techniques for diagnosis to avoid risk of visual impairment. We evaluated two DNA extraction techniques and a semi-nested-PCR (snPCR) targeting the 18S rRNA gene to detect Acanthamoeba cysts and trophozoites. The most effective protocol was evaluated in samples of corneal scrapings and biopsies from an AK rat model and applied to diagnosis of human cases of AK. DNA extraction performed with a commercial kit based on DNA binding to magnetic beads was more efficient than a method based on alkaline lysis, allowing the detection of one trophozoite and one cyst of Acanthamoeba in samples prepared from cultures. This technique and sn-PCR were applied in corneal scrapings of rats experimentally infected with Acanthamoeba (n = 6), resulting in 100% of positivity, against 16.7% (n = 6) of positive identification in culture method using non-nutrient agar (NNA) with Escherichia coli. Corneal biopsies from rats were also tested (n = 6) and resulted in positivity in all samples in both molecular and culture methods. Eight out of ten presumptive human cases of Acanthamoeba keratitis were also confirmed by sn-PCR of scrapping samples, while the culture method was positive in only four cases. We discuss that animal model of AK can be an efficient tool to validate diagnostic methods and conclude that DNA extraction with the kit and snPCR protocol described here is an effective alternative for diagnosis of AK.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acanthamoeba keratitis (AK) is a progressive corneal infection that can cause irreversible vision impairment. The illness is characterized by severe pain, usually disproportionate to the clinical signs, punctate keratopathy, tearing, photophobia, and stromal ring infiltrates (Maycock et al. 2016). Contact lenses users are more likely to acquire AK, representing 62 to 71% of cases (Radford et al. 2002). Corneal traumatic lesions caused by a foreign body followed by exposure to contaminated water can also be a source of AK (Marciano-Cabral and Cabral 2003).
AK requires early diagnosis for a rapid therapeutic intervention, since the prognosis worsens if an effective therapy is not introduced within 3 weeks (Claerhout et al. 2004). History of contact lens use, ocular trauma, or unsuccessful treatments for other disorders such as herpes simplex virus and fungal infections should suggest the possibility of AK (Bacon et al. 1993; Sharma et al. 2000; Sun et al. 2006). Thus, laboratory diagnosis is necessary to confirm the infection. Culture of corneal scrapings is the gold standard for definitive diagnosis of AK (Schuster and Visvesvara 2004) and is also simple and of low cost. However, culture results can take 1 to 10 days, delaying initiation of adequate treatment (Khairnar et al. 2011). Besides, its sensitivity is not high, being reported to range from 7% (Yera et al. 2007) to 57% (Dart et al. 2009).
Polymerase chain reaction-based methods can provide a rapid and sensitive mean of detecting Acanthamoeba in corneal scrapings compared to microbiological and culture methods (Schroeder et al. 2001; Yera et al. 2007; Dhivya et al. 2007; Khairnar et al. 2011). Studies have reported limits of detection of five trophozoites (Khan 2001) and one trophozoite (Schroeder et al. 2001) for PCR using regions of the 18S–rRNA gene as a target. Semi-nested PCR (snPCR) developed by Dhivya et al. (2007) has shown higher sensitivity than uniplex PCR. However, strategies for efficient DNA extraction are needed to avoid false negative results, especially for encysted forms of Acanthamoeba also found in corneal tissue, which are particularly resistant to DNA-extraction procedures validated for other microorganisms (Goldschmidt et al. 2008).
This study aimed to evaluate a protocol for molecular detection of Acanthamoeba in samples of cornea for the diagnosis of AK. Initially, two DNA extraction techniques along with snPCR described by Dhivya et al. (2007) were tested for detection of trophozoites and cysts from cultures. The most effective protocol was evaluated in corneal samples from experimentally infected rats and subsequently applied to diagnose presumptive cases of AK.
Materials and methods
Sample preparation
Trophozoites of an Acanthamoeba polyphaga strain (ATCC 30461) cultured at 30 °C in proteose peptone-yeast extract-glucose (PYG) medium (Visvesvara and Balamuth 1975) were used in the experiments. Cysts were obtained after incubating trophozoites in Neff saline (Neff et al. 1964) for 72 h at 30 °C.
Trophozoites and cysts were quantified in a Neubauer counting chamber, and microtubes containing 50 μL of ultrapure water were inoculated with 1, 5, 10, or 100 cells, in triplicate. Lower quantities (1, 5, and 10) were prepared by microscopic examination of drops of 3–5 μL of diluted suspension on a microscope slide. After identifying the desired quantities, the drop was aspirated and transferred to the microtube. Drops without trophozoites or cysts were also transferred to microtubes to be used as negative controls.
DNA extraction methods
Two protocols of DNA extraction were tested: (1) alkaline lysis and precipitation with ammonium acetate, based on a technique used for DNA extraction of Entamoeba histolytica cysts in feces (Vianna et al. 2009), and (2) a method using the ChargeSwitch® gDNA Mini Tissue Kit (Invitrogen), which is based on DNA binding to magnetic beads.
In the alkaline lysis method, 200 μL of lysis buffer (50 mM glucose, 25 mM Tris HCl pH 8, 10 mM EDTA) and 400 μL of 0.2 mM NaOH with 1% SDS were added to the sample and subjected to seven freeze/thaw cycles (5 min in liquid nitrogen and 5 min at 99 °C). After addition of 300 μL of 7.5 mM ammonium acetate, samples were homogenized and centrifuged at 10,000×g for 5 min. About 700 μL of the supernatant was transferred to a microtube, and 500 μL of isopropanol was added. The mixture was gently homogenized, incubated at −20 °C for 15 min, and centrifuged at 10,000×g for 15 min. The supernatant was discarded and the pellet washed in 70% ethanol. After drying the pellet at ambient temperature, DNA was re-suspended with 25 μL ultrapure water.
For the DNA extraction using the kit, samples were subjected to seven freeze/thaw cycles (5 min in liquid nitrogen and 2 min at 37 °C) in 500 μL of lysis buffer provided in the kit. The subsequent steps of extraction were performed as recommended by the manufacturer’s protocol.
DNA samples from microorganisms known to cause corneal infection were also used to evaluate specificity (Candida albicans, Fusarium solani, Aspergilus fumigatus, Staphylococcus aureus, Pseudomonas aeruginosa, Herpes simplex virus type 1).
Amplification reactions
Briefly, the first round of PCR used the primers JDP1 and JDP2 (Schroeder et al. 2001) in reactions performed on a final volume of 15 μL with 4 mM MgCl2, 0.2 mM dNTP, 0.6 mg/μL bovine serum albumin (BSA), 0.5 μM of each primer, 0.6 U of Platinum® Taq DNA Polymerase (Invitrogen), and 5 μL of DNA. The amplification profile was 94 °C for 7 min, 72 °C for 1 min, 25 cycles at 94 °C for 1 min, 60 °C for 1 min, and 72 °C for 2 min followed by a final extension step of 72 °C for 10 min. The snPCR was performed as described by Dhivya et al. (2007) with minor modifications. Two microliters of PCR product was used for the snPCR carried out on 15 μL of reaction containing 1.6 mM MgCl2, 0.2 mM dNTP, 0.5 μM of the inner forward primer A1 and the outer reverse primer JDP2, and 0.5 U of Platinum® Taq DNA Polymerase (Invitrogen). Amplification was performed at 95 °C for 5 min, followed by 20 cycles at 95, 64, and 72 °C for 45 s each and a final extension at 72 °C for 5 min. All amplification reactions were performed in a thermocycler Biocycler, MJ96 (Biosystems®). The products of snPCR were separated by electrophoresis in 1.5% agarose gel stained with ethidium bromide.
Experimental keratitis in rats
Six female Wistar rats approximately 60 days old were anesthetized with 60 mg/kg ketamine HCl (Vetanarcol®, König do Brasil, Brazil) and 7.5 mg/kg xylazine (Dopaser®, Calier, Spain) by intraperitoneal injection. The left eye was washed three times with sterilized saline solution and then anesthetized with topical 2% lidocaine. Intrastromal inoculation was performed with 2 μL of suspension containing 105 trophozoites of the Acanthamoeba polyphaga ATCC 30461 in Page’s saline (2.5 mM NaCl, 1 mM KH2PO4, 0.5 mM Na2HPO4, 40 mM CaCl2, and 20 mM MgSO4), using a microliter 30G needle, similar to the procedures of Ren and Wu (2010). Two animals were inoculated with Page’s saline only as controls. Six days postinoculation, the lesions were evaluated and the rats euthanized with a double dose of anesthetic before performing corneal scrapings and biopsy. The lesions were graded as follows: Grade I, point of opacity limited to the region of inoculum, less than 1 mm in diameter; Grade II, point of opacity greater than 1 mm, clearly surpassing the initial point of inoculum, but not occupying the entire cornea; and Grade III, extensive damaged area, involving more than 50% of the cornea.
Detection of Acanthamoeba in rat corneal tissue
Each rat inoculated with Acanthamoeba was positioned under a stereomicroscope; the head was immobilized with tape, and the eye washed with sterilized saline. Using a #15 scalpel blade, the injured area of the cornea was scraped three times at paralleled sites at the lesion. Each corneal scraping was inoculated in (1) a tube with 2 mL of PYG medium, (2) a plate non-nutrient agar (NNA) with heated-inactivated Escherichia coli, and (3) a microtube containing 50 μL of Page’s saline. The scalpel was flame-sterilized between each procedure. For biopsy, a 3-mm fragment of the cornea, incorporating both corneal infiltrate and adjacent clear cornea, was excised and removed with a fine tip tweezers to a sterilized lamina. The fragment was separated into three parts for culture in PYG, culture in NNA, and inoculation in a microtube with Page’s saline, as previously described for scrapings. Corneas inoculated only with Page’s saline were submitted to the same procedures as controls. Cultures were incubated at 30 °C and examined from day 5 to day 20 following the seeding. Samples collected in the microtubes with 50 μL of Page’s saline were stored at −20 °C until DNA extraction using ChargeSwitch® gDNA Mini Tissue Kit (Invitrogen) and snPCR.
Diagnosis of human cases of Acanthamoeba keratitis
Ten presumptive human cases of AK admitted in Ophtalmology Sector of Cassiano Antônio de Moraes Universitary Hospital, Vitória, Espírito Santo, Brazil, during the period of March 2010 and May 2012, were evaluated by culture and PCR/snPCR. Corneal samples were seeded in soy-agar plates with Escherichia coli, as described by Costa et al. (2010). A second scraping was transferred to microtubes with 200 μL de Page’s saline and immediately transported to the laboratory for centrifugation at 5000×g by 5 min. About 150 μL of the supernatant was discarded and the microtube was stocked at −20 °C until performing DNA extraction, which was carried out with ChargeSwitch® gDNA Mini Tissue Kit (Invitrogen).
Results
The snPCR has detected one trophozoite in two out of three repetitions using both DNA extraction methods (Table 1). When cysts were used for DNA extraction, positivity by snPCR was achieved with 100 forms treated by alkaline lysis extraction and 1 to 10 forms subjected to the commercial kit (although this did not occur in all repetitions) (Table 1). The samples made of ultrapure water (negative controls) and DNA samples obtained from fungi, bacteria, and virus (specificity controls) had a negative result in the tests.
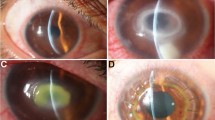
Keratitis induced in Wistar rats resulted in corneal lesions from grades I to III in all animals (Fig. 1, Table 2). Corneal inoculation with Page’s saline resulted in no lesions. Table 2 outlines results of the cultures and snPCR, using corneal scraping or corneal biopsy from the animals. For corneal scraping samples, culture in PYG was negative, culture in NNA was positive in only one sample, and snPCR showed signs of amplification in all six samples. In corneal biopsy material, all six samples were positive in NNA cultures, while four samples were detected by culture in PYG. All corneal biopsy samples were positive for Acanthamoeba by snPCR. Samples from rat cornea inoculated with Page’s saline were negative in all diagnostic methods.
Acanthamoeba keratitis induced in Wistar rats by intrastromal inoculation of 105 trophozoites of A. polyphaga strain ATCC 30461. Lesions were evaluated 5 days after the inoculation. Lesions classified as grade II, with the point of opacity greater than 1 mm, clearly exceeding the initial point of inoculum (a) and grade III, showing an extensive damaged area and involvement of more than 50% of the cornea (b)
Four out of ten corneal scrapings of presumptive cases of Acanthamoeba keratitis were positive in culture method and eight were positive in snPCR (Table 3).
Discussion
In recent years, both conventional PCR and real-time PCR have been reported to be efficient techniques to detect Acanthamoeba in corneal tissue (Thompson et al. 2008; Khairnar et al. 2011; Laummaunwai et al. 2012). Conventional PCR is widespread, and lower cost equipments and reagents have recently become available. However, limitations such as the low number of amoebae in the corneal scrapings and the greater resistance of cysts to extraction and release of DNA must be considered in establishing a protocol using molecular techniques. In the present study, we have evaluated a DNA extraction method that was efficient to detect one trophozoite and one cyst in snPCR. We have also validated it in corneal samples of animal and, subsequently, in human scrapings.
Trophozoites and cysts from cultures were used in the initial experiments to determine an efficient DNA extraction technique, including the evaluation of a protocol of alkaline lysis to obtain DNA from Entamoeba histolytica and E. dispar in feces (Vianna et al. 2009). The protocol was adapted and tested on the assumption that lysis with NaOH could facilitate breaking the cyst wall and releasing the DNA from cysts. The method seems to be relatively efficient for trophozoites, since two out of three repetitions using one and five cells resulted in positivity with snPCR. However, no sign of amplification was observed in snPCR when 10 or fewer cysts were used. These results confirmed the already known resistance of Acanthamoeba cysts to DNA extraction, which seems to be related to the resistance of the cyst wall as well as the presence of high-density bodies in the cyst nucleus. These bodies are consisted of core protein condensed by genetic material that can interfere with the DNA denaturation (Goldschmidt et al. 2008; Lasman 1977).
When the extraction kit was used, it was possible to detect 1, 5, and 10 trophozoites or cysts by snPCR in one or more of the triplicate samples. A possible explanation for the higher efficiency of the kit is the use of proteinase K, which could increase the DNA available for the binding to primers and for polymerase action (Goldschmidt et al. 2008). The possibility of false-positive detection as a result of contamination (amplicons) was prevented by careful handling during the procedures and monitoring with control samples, which were all negative.
To obtain corneal scrapings and biopsy for evaluating the molecular technique and to compare it to culture methods, Acanthamoeba keratitis was induced in a rat model. The intrastromal inoculation resulted in lesions of variable grades in all the six animals tested. Although intrastromal inoculation did not replicate the most frequent source of contamination in humans, it provided a higher infection rate, compared to procedures like corneal scratching and the wearing of contaminated contact lenses (Ren and Wu 2010). Thus, the rat model for keratitis could be a good alternative for testing diagnostic procedures applicable to human patients.
Culture in agar plates with Escherichia coli is the usual method of diagnosis of Acanthamoeba in corneal samples (Siddiqui and Khan 2012). In this study, this method was compared to seeding in PYG medium and seemed to be slightly superior for diagnostic purposes. As the agar plate method is simpler and uses a lower number of components, it was considered the best choice in terms cost-benefit. Furthermore, the higher number of nutrients in PYG could, in some instances, stimulate the growth of organisms other than Acanthamoeba in coinfection cases, masking the results.
Comparing the corneal materials, biopsy was clearly superior to corneal scraping regarding the positivity in culture, which can be explained by the higher number of parasites obtained in biopsy. It is known that biopsy is the indicated procedure in cases when the microbial agent is located in deeper layers of the cornea or when corneal scrapings are repeatedly negative, since scraping can be insufficient for the isolation (Younger et al. 2012). However, corneal scraping is the chosen procedure in suspected cases of amoebic keratitis, since it is less invasive and easier to perform. In this study, the snPCR was efficient to detect Acanthamoeba in corneal scraping in the animal model, showing that a small amount of tissue can be a limitation for culturing, but not for snPCR. Evaluation of human corneal scrapings for diagnosis of Acanthamoeba keratitis also confirmed this point. The human samples that were negative in snPCR were also negative in the culture, suggesting that these keratitis cases should be caused by other causes rather than Acanthamoeba. Indeed, empiric treatment for Acanthamoeba did not result in improvement of the symptoms, and other causes were then attributed for these cases (data not shown).
In conclusion, DNA extraction with Charge Switch® Kit after freeze/thaw process and amplification by snPCR presented here was efficient to detect the low quantities of trophozoites and cysts that may be present in corneal scrapings. The use of the technique in corneal samples of infected animals and its validation to diagnose human-suspected cases indicated that this is a good alternative for diagnosis of Acanthamoeba keratitis.
References
Bacon AS, Frazer DG, Dart JKG, Matheson M, Ficker LA, Wright P (1993) A review of 72 consecutive cases of Acanthamoeba keratitis, 1984–1992. Eye 7:719–725. doi:10.1038/eye.1993.168
Claerhout I, Goegebuer A, Van Den Broecke C, Kestelyn P (2004) Delay in diagnosis and outcome of Acanthamoeba keratitis. Graefes Arch Clin Exp Ophthalmol 242:648–653. doi:10.1007/s00417-003-0805-7
Costa AO, Castro EA, Ferreira GA, Furst C, Crozeta MA, Thomaz-Soccol V (2010) Characterization of Acanthamoeba isolates from dust of a public hospital in Curitiba, Paraná, Brazil. J Eukaryot Microbiol 57:70–75. doi:10.1111/j.1550-7408.2009.00453.x
Dart JKG, Saw VPJ, Kilvington S (2009) Acanthamoeba keratitis: diagnosis and treatment update 2009. Am J Ophthalmol 148:487–499.e2. doi:10.1016/j.ajo.2009.06.009
Dhivya S, Madhavan HN, Rao CM, Ramchander PV, Therese KL, Malathi J (2007) Comparison of a novel semi-nested polymerase chain reaction (PCR) with a uniplex PCR for the detection of Acanthamoeba genome in corneal scrapings. Parasitol Res 100:1303–1309. doi:10.1007/s00436-006-0413-7
Goldschmidt P, Degorge S, Saint-Jean C, Yera H, Zekhnini F, Batellier L, Laroche L, Chaumeil C (2008) Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br J Ophthalmol 92:112–115. doi:10.1136/bjo.2007.125898
Khairnar K, Tamber GS, Ralevski F, Pilla DR (2011) Comparison of molecular diagnostic methods for the detection of Acanthamoeba spp. from clinical specimens submitted for keratitis. Diagn Microbiol Infect Dis 70:499–506. doi:10.1016/j.diagmicrobio.2011.03.019
Khan NA (2001) Pathogenicity, morphology, and differentiation of Acanthamoeba. Curr Microbiol 43:391–395. doi:10.1007/s002840010325
Lasman M (1977) Light and electron microscopic observations on encystment of Acanthamoeba palestinensis, Reich. J Protozool 24:244–248
Laummaunwai P, Ruangjirachuporn W, Boonmars T (2012) A simple PCR condition for detection of a single cyst of Acanthamoeba species. Parasitol Res 110:1569–1572. doi:10.1007/s00436-011-2662-3
Marciano-Cabral F, Cabral G (2003) Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16:273–307. doi:10.1128/CMR.16.2.273-307.2003
Maycock NJR, Jayaswal R, Ed F (2016) Update on Acanthamoeba Keratitis: diagnosis, treatment and outcomes. Cornea 35:713–720. doi:10.1097/ICO.0000000000000804
Neff R, Ray S, Benton W, Wilborn M (1964) Induction of synchronous encystment (differentiation) in Acanthamoeba sp. Methods Cell Physiol 55–83
Radford CF, Minassian DC, Dart JKG (2002) Acanthamoeba keratitis in England and Wales: incidence, outcome, and risk factors. Br J Ophthalmol 86:536–542
Ren M, Wu X (2010) Evaluation of three different methods to establish animal models of Acanthamoeba keratitis. Yonsei Med J 51:121–127. doi:10.3349/ymj.2010.51.1.121
Schroeder JM, Booton GC, Hay J, Niszl IA, Seal DV, Markus MB, Fuerst PA, Byers TJ (2001) Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol 39:1903–1911. doi:10.1128/JCM.39.5.1903-1911.2001
Schuster FL, Visvesvara GS (2004) Amebae and ciliated protozoa as causal agents of waterborne zoonotic disease. Vet Parasitol 126:91–120. doi:10.1016/j.vetpar.2004.09.019
Sharma S, Garg P, Rao GN (2000) Patient characteristics, diagnosis, and treatment of non-contact lens related Acanthamoeba keratitis. Br J Ophthalmol 84:1103–1108
Siddiqui, Khan (2012) Biology and pathogenesis of Acanthamoeba. Parasites & Vectors 2012 5:6. doi:10.1186/1756-3305-5-6
Sun X, Zhang Y, Li R, Wang Z, Luo S, Gao M, Deng S, Chen W, Jin X (2006) Acanthamoeba keratitis: clinical characteristics and management. Ophthalmology 113:412–416. doi:10.1016/j.ophtha.2005.10.041
Thompson PP, Kowalski RP, Shanks RMQ, Gordon YJ (2008) Validation of real-time PCR for laboratory diagnosis of Acanthamoeba keratitis. J Clin Microbiol 46:3232–3236. doi:10.1128/JCM.00908-08
Vianna EN, Costa JO, Santos CKS, Cury MC, Silva EF, Costa AO, Gomes MA (2009) An alternative method for DNA extraction and PCR identification of Entamoeba histolytica and E. dispar in fecal samples. Parasitology 136:765. doi:10.1017/S0031182009006167
Visvesvara GS, Balamuth W (1975) Comparative studies on related free-living and pathogenic amebae with special reference to Acanthamoeba. J Protozool 22:245–256
Yera H, Zamfir O, Bourcier T, Viscogliosi E, Noel C, Dupouy-Camet J, Chaumeil C (2007) Comparison of PCR, microscopic examination and culture for the early diagnosis and characterization of Acanthamoeba isolates from ocular infections. Eur J Clin Microbiol Infect Dis 26:221–224. doi:10.1007/s10096-007-0268-6
Younger JR, Johnson RD, Holland GN, Page JP, Nepomuceno RL, Glasgow BJ, Aldave AJ, Yu F, Litak J, Mondino BJ, UCLA Cornea Service (2012) Microbiologic and histopathologic assessment of corneal biopsies in the evaluation of microbial keratitis. Am J Ophthalmol 154:512–519.e2. doi:10.1016/j.ajo.2012.03.014
Acknowledgements
The authors thank CNPq, Fundação Araucária, and PRPq/UFMG for financial support. We also thank Ophthalmology Sector staff from Cassiano Antônio de Moraes University Hospital for providing the human corneal scrapings used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures with animal were conducted under accepted guidelines for the care and use of laboratory animals for research and approved by the Ethical Committee for Animal Experimentation of the Biological Sciences Sector of Federal University of Paraná, Brazil, protocol number 420b.
The use of information on the diagnosis of patients suspected of Acanthamoeba keratitis was approved by the Ethical Committee of Health Sciences Center of Federal University of Espírito Santo, Vitória, ES, Brazil (protocol number 006/07), and followed the tenets of Declaration of Helsinki.
Rights and permissions
About this article
Cite this article
Costa, A.O., Furst, C., Rocha, L.O. et al. Molecular diagnosis of Acanthamoeba keratitis: evaluation in rat model and application in suspected human cases. Parasitol Res 116, 1339–1344 (2017). https://doi.org/10.1007/s00436-017-5411-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5411-4