Abstract
The aim of the study was to identify the Cryptosporidium parvum subtypes circulating in Polish cattle and their distribution in relation to the age and health status of tested animals. In total, 779 fecal samples were obtained from young cattle originating from 237 farms. C. parvum strains were identified at the 18 small-subunit ribosomal RNA (SSU rRNA), COWP, and LIB13 loci and were subsequently analyzed by sequencing at the 60-kDa glycoprotein (GP60) locus for subtype determination. The presence of 71 C. parvum strains belonging to IIa, IId, or IIl subtype families was shown. The strains from the IIa allele family prevailed with IIaA17G1R1, IIaA17G2R1, and IIaA15G2R1 subtypes occurring frequently. Two novel subtypes IIaA10G1R1 and IIlA19R3 were detected for the first time in a bovine host. The highest C. parvum prevalence (22.5 %, confidence interval (CI) = 2.5 %) was observed among the youngest animals up to 2 weeks of age, followed by the prevalence among those aged 2 to 4 weeks (6.6 %, CI = 2.6 %) and then among older cattle (4.9 %, CI = 2.1). The occurrence of diarrhea in animals was associated with the presence of the IIaA16G1R1b subtype, while infections caused by IIaA15G2R1 strains were more likely to be asymptomatic. The geographical distribution of subtypes revealed that strains from the IIa subtype family were detected all over the country frequently compared to the IId and IIl subtypes, the sporadic appearances of which confirmed their endemic occurrence. Subtype analysis revealed the presence of zoonotic strains indicating cattle as a reservoir for human cryptosporidiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium infections are often reported in cattle raised in different geographical regions under various husbandry systems (de Graaf et al. 1999). Despite observed worldwide prevalence of the parasite in different animal species, infections have usually generated considerable losses in the livestock industry (Olson et al. 2004). Calves acquire infections shortly after birth, and animals below the age of 1 month are the major host of the parasite (Santín and Trout 2008; Rzeżutka and Kaupke 2013). Cattle can be infected by several Cryptosporidium species, but only two of these cattle-associated species, Cryptosporidium parvum and Cryptosporidium andersoni, have been found to cause human cryptosporidiosis (Leoni et al. 2006; Liu et al. 2014). It has previously been shown that the bovine host is a major reservoir of zoonotic C. parvum and contact with an infected animal or drinking contaminated water can lead to human infection (Hunter and Thompson 2005; Cacciò et al. 2005; Olson et al. 2004). Sequencing of a 60-kDa glycoprotein (GP60) gene is a frequently used subtyping method (Waldron et al. 2009; Plutzer and Karanis 2007; Díaz et al. 2012). This tool also confirmed its usefulness during epidemiological investigation of cryptosporidiosis cases and in surveillance studies of human and animal cryptosporidiosis (Soba and Logar 2008). Based on sequence analysis of the GP60 gene, the existence of 14 (IIa–IIo) C. parvum subtype families has been shown in humans and animals (Insulander et al. 2013; Wang et al. 2014). The analysis of a microsatellite region within the GP60 gene of C. parvum strains revealed the parasite’s heterogeneity, with a large number of subtypes detected within each subtype family that encompasses zoonotic or human-specific types. Although a lot of studies have been conducted that aimed to subtype C. parvum strains detected in humans and animals, our knowledge on subtype occurrence and worldwide distribution in the human and animal host is still not complete (Xiao 2010). So far, little is known about the occurrence and epidemiology of infections in humans caused by zoonotic C. parvum subtypes in Poland, although information on possible zoonotic transmission (Majewska et al. 1999; Gait et al. 2008) and identification of zoonotic species present in farm animals exists (Rzeżutka and Kaupke 2013; Kaupke et al. 2014; Rzeżutka et al. 2014; Majewska et al. 2000, 2001, 2004). The aim of the study was to identify the C. parvum subtypes circulating in the Polish cattle population, in respect to the presence of zoonotic subtypes. In addition, animal age and animal health as factors in the occurrence and distribution pattern, respectively, of parasite subtypes were investigated.
Materials and methods
Origin of the samples
Fecal samples were collected from calves from birth to 2 months of age originating from 237 farms located in all 16 provinces in Poland. In total, 779 cattle fecal samples were obtained during 4 years of monitoring from 2010 to 2014. Sampled farms and the animals on those farms were randomly selected for the studies. They represented different administrative locations across each province. Average herd size differs in the sampled farms, although the minimum number of heads per farm was 20. From each farm, from two to five individual fecal samples of approximately 10–15 g were placed into plastic containers, labeled, and sent to the laboratory. Of these samples, 633 had previously been tested for Cryptosporidium, resulting in detection of 34 C. parvum strains but without pursuant subtype identification (Rzeżutka and Kaupke 2013). The majority of sampled animals (689 calves) did not show any gastrointestinal disorders, and only 90 calves did, demonstrating diarrheal illness (with loose and watery stools) of an unknown etiology on the day of sampling. The age and health status of sampled cattle are presented in Table 1.
Detection of C. parvum in cattle and subtype identification
Samples were analyzed using molecular methods according to a previously described procedure. Briefly, parasite genomic DNA was extracted from 0.1 g of the feces with an alkali wash and a heat lysis method developed by Millar et al. (2001) with further modifications (Rzeżutka and Kaupke 2013). Identification of C. parvum was performed at the 18 small-subunit ribosomal RNA (SSU rRNA) (Xiao et al. 1999), COWP (Homan et al. 1999), and LIB13 (Tanriverdi et al. 2003) loci by conventional PCR. The LIB13 PCR was a C. parvum and Cryptosporidium hominis-specific assay targeting sequence polymorphism of a four-nucleotide deletion on an unknown genomic sequence of C. parvum. The positive 18 SSU rRNA (849 bp) and COWP (640 bp) PCR products were subjected to restriction fragment length polymorphism (RFLP) analysis using NdeI (Zintl et al. 2007) and TaqI (Homan et al. 1999) enzymes. For subtyping, two nested PCR assays that target the GP60 gene locus giving amplicons of approximately 800 or 400 bp were used (Glaberman et al. 2002; Sulaiman et al. 2005). The appropriate positive and negative controls were included during the nucleic acid extraction and PCR analyses. Visualization of PCR amplicons and digested products was performed by electrophoresis in either 1.7 or 2.5 % agarose gels stained with ethidium bromide. The GP60 amplicons were subsequently excised from the agarose gel, purified, sequenced, and compared with the reference sequences of each subtype family submitted to the GenBank database using the NCBI BLASTn program (http://blast.ncbi.nlm.nih.gov). Particular subtypes were identified based on trinucleotide repeats present in analyzed sequences.
Statistical analysis
Moreover, the relationship between cattle age and frequency of C. parvum occurrence as well as the dominance of infections caused by C. parvum strains from the IIa subtype family over strains belonging to the IId and IIl families were statistically analyzed using one-way analysis of variance (ANOVA) with Tukey confidence intervals (CIs). Subsequently, the relationship between infections caused by a particular subtype and the presence of diarrhea was determined on the basis of the confidence interval for the mean estimated for all diarrheic animals. A chi-square test (χ 2) was used to determine the frequency of C. parvum infections in diarrheic and healthy animals as well as to show the relationship between the animal age and the occurrence of infections caused by various C. parvum subtypes. Concluding the statistical work, a two-way ANOVA without interactions was applied to determine how the relationship between the animal age and the presence of each subtype exerts influence on the frequency of infections caused by these strains. All calculations were performed with Statgraphics Centurion v. XV.
Results
Molecular identification: PCR-RFLP, GP60 sequencing, and sequence analysis
Cryptosporidium detection performed at the 18 SSU rRNA, COWP, and GP60 loci revealed the presence of C. parvum in 76 samples analyzed; however, a successful subtype identification was obtained for 71 strains belonging to the IIa, IId, or IIl subtype families. The IIa subtype family prevailed (84.2 %, CI = 3.6 %) compared to IId (9.5 %, CI = 4.2 %) and IIl (6.0 %, CI = 7.2 %) subtypes. The IIa subtype family was represented by the following subtypes: IIaA17G1R1 (n = 21), IIaA17G2R1 (n = 14), IIaA15G2R1 (n = 14), IIaA16G1R1b (n = 8), IIaA10G1R1 (n = 2), IIaA14G1R1 (n = 2), IIaA16G3R1 (n = 2), IIaA18G1R1c (n = 1), and IIaA19G1R1 (n = 1). The IId subtype family was found in five samples, specifically IIdA23G1 (n = 2), IIdA24G1c (n = 2), and IIdA22G1b (n = 1), and one sample contained IIl with the IIlA19R3 identity. For the first time, two novel subtypes, IIaA10G1R1 and IIlA19R3, were detected in a bovine host. None of the tested samples contained more than one C. parvum GP60 subtype family. PCR amplification with GP60 primers was unsuccessful only for five C. parvum strains, probably either because of the low abundance of Cryptosporidium DNA present or because of heterogeneity within the primer’s sequences to the DNA sequence of the subtype present. Sequences representing each subtype within the tested group of animals were deposited in GenBank (Table 1).
Distribution of C. parvum subtypes in relation to health status and animal age
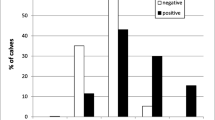
C. parvum infections were significantly more frequently identified (χ 2 = 25.24, P < 0.001) in diarrheic cattle (40 %) than in healthy animals (5.8 %). The frequency of infections between different age groups of calves (birth–2 weeks, >2–4 weeks, and >4–8 weeks) differed significantly (P = 0.022, α = 0.05). Indeed, the highest C. parvum prevalence (22.5 %, CI = 2.5 %) was observed in animals at the age of up to 2 weeks. It was significantly lower for animals at the age between 2 and 4 weeks (6.6 %, CI = 2.6 %) and older (4.9 %, CI = 2.1 %) (Fig. 1).
Calves were mostly infected with four out of 14 subtypes detected (IIaA17G1R1, IIaA17G2R1, IIaA15G2R1, and IIaA16G1R1b). The part of diarrheic animals in all cattle infected by these subtypes was 57.74 ± 37.63 %. The designated confidence interval for the mean (20.11–95.37) shows that the infections caused by IIaA15G2R1 were more likely to be asymptomatic (7.1 < 20.11 %), while the presence of IIaA16G1R1b subtypes was always associated with diarrhea (100 > 95.37 %) (Fig. 2). Of note, these animals were kept on two farms situated in different areas in Wielkopolska Province. For IIaA17G1R1 and IIaA17G2R1 subtypes, there was no relationship observed between the presence and absence of diarrhea during the course of infection. Nevertheless, it should be noticed that animals infected by these subtypes are more likely to develop diarrhea (values of 66.7 and 57.1 % are within the confidence interval for the mean).
There was no correlation observed between the cattle age and the presence of a particular subtype (P = 0.869, α > 0.05). Also, there was no statistically significant relationship present between the animal age (P = 0.452, α > 0.05), the presence of specific subtype (P = 0.213, α > 0.05), and the frequency of infections caused by them. Nevertheless, particular subtypes were only shown in animals at a certain age. For example, IIaA10G1R1, IIaA14G1R1, IIaA19G1R1, IIdA22G1b, and IIdA24G1c subtypes were detected in the youngest animals (below 2 weeks of age), while IIdA23G1 and IIlA19R3 subtypes occurred in cattle at the age between 2 and 4 weeks, being followed by the IIaA18G1R1c subtype found solely in cattle older than 4 weeks (Fig. 3).
Geographical distribution of subtypes
The infected animals were detected on 44 (18.6 %) of the 237 monitored farms. Among the three commonly detected IIa subtypes, IIaA17G1R1 was found in six out of 16 Polish administrative provinces. In comparison to subtypes from the IId and IIl allele families, strains from the IIa subtype family were frequently detected all over the country (Fig. 4). They were present in 39 (16.4 %) of the investigated farms. The IId subtypes were found sporadically in four farms localized in Lublin, Pomerania, and West Pomerania provinces. The animal harboring IIlA19R3 was raised in Lublin Province.
Discussion
Genome sequencing and multilocus analysis of highly polymorphic sequences have significantly increased subtyping resolution and knowledge of the genetic structure of Cryptosporidium (Xiao 2010). In this study, subtyping of C. parvum strains was performed based on the DNA sequence polymorphism analysis of the GP60 gene. Cryptosporidium infections have been reported in cattle worldwide with a varied prevalence regardless of the animal age ranging from 3.5 % (Epe et al. 2004) to 60.2 % (Mišic and Abe 2007). In the studies presented, C. parvum prevalence was estimated at 9.7 % only for animals up to the age of 2 months. This prevalence is not different from that reported by other authors in cattle being at the same age (Brook et al. 2009; Björkman et al. 2015). Nevertheless, only 18.6 % of farms were positive for this parasite species, indicating its low occurrence on the farm level. The differences observed between farms could be associated with the age of animals sampled, as the prevalence is age related and is higher in younger animals and varied geographical occurrence and distribution across the sampled regions; therefore, studies limited only to restricted area will not give a good estimation for parasite prevalence. Other factors that have an influence on prevalence are the herd size, type of breed, farm management, or husbandry system.
It was shown that the majority (91.5 %) of identified C. parvum strains in cattle belonged to the IIa zoonotic allele family. There were also infections detected which were caused by IId (7.04 %) and, rare for Europe, IIl (1.4 %) subtype families. It was statistically confirmed that the IIa subtypes prevailed in cattle regardless of animal age or geographical location of the farm. Subtype analysis revealed that the IIaA17G1R1 isolate was mainly responsible for the infection, as it was present in 29.6 % of C. parvum-positive samples. This dominating subtype in Polish cattle has previously been found in sheep (Plutzer and Karanis 2007; Wielinga et al. 2008; Thompson et al. 2007; Stantič-Pavlinic et al. 2003) and in cattle with prevalence among tested animals ranging from 53.3 to 98.4 % (Geurden et al. 2007; Brook et al. 2009; Duranti et al. 2009; Broglia et al. 2008; Wielinga et al. 2007; Alves et al. 2003, 2006; Imre et al. 2009, 2010; Soba and Logar 2008; Quilez et al. 2008; Rieux et al. 2013). Apart from the dominating IIaA17G1R1 isolates, IIaA15G2R1 and IIaA17G2R1 subtypes also frequently occurred. It is worth noting that eight (A14G1R1, A15G2R1, A16G1R1b, A16G3R1, A17G1R1, A17G2R1, A18G1R1c, and A19G1R1) out of nine IIa subtypes identified in this study have previously been described as cattle pathogens (Thompson et al. 2007; Brook et al. 2009; Mišic and Abe 2007; Silverlås et al. 2013). Furthermore, concurrent infections caused by more than one subtype were not reported in any animal. It was shown that the frequency of C. parvum infections in cattle is age related, with the highest occurrence of this parasite in animals at the age of 1 day to 2 weeks. For frequently appearing subtypes (IIaA15G2R1, IIaA16G1R1b, IIaA17G1R1, and IIaA17G2R1), the relationship between the occurrence of subtypes and its higher pathogenicity as manifested by cattle diarrhea was investigated. In fact, this association was only shown for the IIaA16G1R1b subtype as diarrhea was attributed to all infections caused by this strain. Profuse diarrhea was also seen in one calf harboring the IIlA19R3 subtype. Despite treatment, this animal died from a diarrheal disease, and subsequent laboratory investigation excluded bacteria as a primary cause of disease (personal communication). However, when the course of the infection was asymptomatic, it was associated with the presence of IIaA15G2R1. The relationship between the presence of any specific C. hominis or C. parvum subtype and the occurrence of clinical symptoms was also investigated in humans by Cama et al. (2007) and in calves by Geurden et al. (2007). However, in the case of C. parvum subtypes, the existence of such a congruity was not confirmed. In contrast to our findings, the IIaA15G2R1 strain was found not only in diarrheic but also in non-diarrheic dairy calves in France (Rieux et al. 2013). Nevertheless, the relationship between clinical signs and subtype occurrence was not analyzed statistically.
In this study, four different subtypes comprising IIdA22G1b, IIdA23G1, IIdA24G1c, and IIlA19R3 from the less common IId and IIl genetic families were detected. Some of these subtypes have previously been found in cattle in Spain and Sweden (Quilez et al. 2008; Silverlås et al. 2010) with the high natural prevalence of IId strains reported in cattle housed outside Europe (Amer et al. 2010; Muhid et al. 2011; Zhang et al. 2013). In contrast to IIa and IId subtypes, isolates from the IIl genetic lineage have also occasionally been reported in Europe (Soba and Logar 2008; Wielinga et al. 2008). In this study, there were no unusual subtypes detected except one IIaA10G1R1, identified in two asymptomatic animals kept on the same farm. This is the first report describing its presence in cattle. However, based on the existing data, it is difficult to determine its significance in the epidemiology of bovine cryptosporidiosis. There was no correlation observed between strain distribution and region of the country, except in the case of isolates from the IId and IIl allele families present solely in north and east Poland. When a subtype distribution is compared with this observed in neighboring countries, the dominating subtype (IIaA17G1R1) in Poland has not been found in cattle neither in Germany nor in the Czech Republic. Surprisingly, the IIaA15G2R1 subtype prevailing in Germany has also wide distribution in the west and central part of Poland.
Genetic diversity of detected strains within the same farm was not observed, suggesting endemicity of a single subtype on a farm. As seen in recent studies, this finding cannot be considered as a rule (Abeywardena et al. 2012); however, similar, low variation among subtypes detected in animals housed on the same farm has been described in Slovenia, Serbia, Montenegro, and France (Mišic and Abe 2007; Soba and Logar 2008; Rieux et al. 2013). Certainly, greater genetic variability within the analyzed population of C. parvum strains might be found if multilocus analyses targeting mini- and microsatellite loci were analyzed. The association between the animal age, the occurrence of particular subtype, and the frequency of infections was not confirmed statistically. The lack of interaction in the statistical model resulted from non-orthogonality of data, i.e., the presence of different subtypes and their presence in varying numbers in each group of tested animals.
It is known that cryptosporidiosis in humans is not mainly caused by C. hominis, but rather that other Cryptosporidium species present in livestock and free-living animals represent more important human pathogens and a common cause of diarrhea (Ryan and Hijjawi 2015). Although C. parvum subtypes from IIa and IId allele families are considered to be zoonotic, not all currently identified members of these families have been detected in humans. This data may partly support assumptions that animals could be an exclusive reservoir for some subtypes (Abe et al. 2006; Wielinga et al. 2007; Soba and Logar 2008; Hijjawi et al. 2010; O’Brien et al. 2008; Waldron et al. 2009; Budu-Amoako et al. 2012). Nevertheless, the zoonotic nature of some C. parvum strains detected in this study has previously been confirmed (Soba and Logar 2008; Hijjawi et al. 2010; Chalmers et al. 2011; Lassen et al. 2014) with the IIaA15G2R1 subtype often being detected in both humans and animals (Chalmers et al. 2011; Drumo et al. 2012). In fact, cases of human cryptosporidiosis in Poland are only sporadically identified and typically without subsequent information on disease-causing subtypes. This lack of data significantly hinders an epidemiological investigation aiming at recognition of the infection source. Although the studies were conducted on randomly selected animals originated from farms at different locations, the number of cattle tested from each province did not reflect the size of population being kept in this region. It could be taken as a major limitation in the interpretation of results on the occurrence and prevalence of C. parvum subtypes in Polish cattle. In addition, an attempt was undertaken to investigate the relationship between infections caused by particular subtypes and the presence of diarrhea in infected animals. Although some relationships were shown and confirmed statistically, the low number of strains representing rarely detected subtypes did not allow to fully investigate their clinical significance. Cryptosporidium diarrhea in calves is often associated with C. parvum infections compared to other parasite species frequently detected in this type of host. Because the etiology of diarrhea in calves was not thoroughly investigated for the possible co-infections with bacterial and viral pathogens, therefore, its strict association solely to Cryptosporidium infection might tend to overestimate some findings.
References
Abe N, Makoto M, Isao K, Iseki M (2006) Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res 99:303–305
Abeywardena H, Jex AR, Nolan MJ, Haydon SR, Stevens MA, McAnulty RW, Gasser RB (2012) Genetic characterisation of Cryptosporidium and Giardia from dairy calves: discovery of species/genotypes consistent with those found in humans. Infect Genet Evol 12:1984–1993
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41:2744–2747
Alves M, Xiao L, Antunes F, Matos O (2006) Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol Res 99:287–292
Amer S, Harfoush M, He H (2010) Molecular and phylogenetic analyses of Cryptosporidium spp from dairy cattle in Egypt. J Egypt Soc Parasitol 40:349–366
Björkman C, Lindström L, Oweson C, Ahola H, Troell K, Axén C (2015) Cryptosporidium infections in suckler herd beef calves. Parasitology 142:1108–1114
Broglia A, Reckinger S, Cacció SM, Nöckler K (2008) Distribution of Cryptosporidium parvum subtypes in calves in Germany. Vet Parasitol 154:8–13
Brook EJ, Anthony Hart C, French NP, Christley RM (2009) Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Vet J 179:378–382
Budu-Amoako E, Greenwood SJ, Dixon BR, Barkema HW, McClure JT (2012) Occurrence of Cryptosporidium and Giardia on beef farms and water sources within the vicinity of the farms on Prince Edward Island, Canada. Vet Parasitol 184:1–9
Cacciò SM, Thompson RC, McLauchlin J, Smith HV (2005) Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol 21:430–437
Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L (2007) Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis 196:68
Chalmers RM, Smith RP, Hadfield SJ, Elwin K, Giles M (2011) Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasitol Res 108:1321–1325
de Graaf DC, Vanopdenbosch E, Ortega-Mora LM, Abbassi H, Peeters JE (1999) A review of the importance of cryptosporidiosis in farm animals. Int J Parasitol 29:1269–1287
Díaz P, Hadfield SJ, Quílez J, Soilán M, López C, Panadero R, Díez-Baños P, Morrondo P, Chalmers RM (2012) Assessment of three methods for multilocus fragment typing of Cryptosporidium parvum from domestic ruminants in north west Spain. Vet Parasitol 186:188–195
Drumo R, Widmer G, Morrison LJ, Tait A, Grelloni V, D’Avino N, Pozio E, Cacciò SM (2012) Evidence of host-associated populations of Cryptosporidium parvum in Italy. Appl Environ Microbiol 78:3523–3529
Duranti A, Cacciò SM, Pozio E, Di Egidio A, De Curtis M, Battisti A, Scaramozzino P (2009) Risk factors associated with Cryptosporidium parvum infection in cattle. Zoonoses Public Health 56:176–182
Epe C, Coati N, Schnieder T (2004) Results of parasitological examinations of faecal samples from horses, ruminants, pigs, dogs, cats, hedgehogs and rabbits between 1998 and 2002. Dtsch Tierarztl Wochenschr 111:243–247
Gait R, Soutar RH, Hanson M, Fraser C, Chalmers R (2008) Outbreak of cryptosporidiosis among veterinary students. Vet Rec 162:843–845
Geurden T, Berkvens D, Martens C, Casaert S, Vercruysse J, Claerebout E (2007) Molecular epidemiology with subtype analysis of Cryptosporidium in calves in Belgium. Parasitology 134:1981–1987
Glaberman S, Moore JE, Lowery CJ, Chalmers RM, Sulaiman I, Elwin K, Rooney PJ, Millar BC, Dooley JS, Lal AA, Xiao L (2002) Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg Infect Dis 8:631–633
Hijjawi N, Ng J, Yang R, Atoum MF, Ryan U (2010) Identification of rare and novel Cryptosporidium GP60 subtypes in human isolates from Jordan. Exp Parasitol 125:161–164
Homan W, van Gorkom T, Kan YY, Hepener J (1999) Characterization of Cryptosporidium parvum in human and animal feces by single-tube nested polymerase chain reaction and restriction analysis. Parasitol Res 85:707–712
Hunter PR, Thompson RC (2005) The zoonotic transmission of Giardia and Cryptosporidium. Int J Parasitol 35:1181–1190
Imre K, Matos O, Dărăbus G, Mederle N, Oprescu I, Morariu S, Ilie MS, Hotea I, Imre M (2009) First genetic identification of Cryptosporidium spp. in cattle in Romania. Lucr Şt Med Vet Timişoara 52:26–30
Imre K, Dărăbuş G, Mederle N, Oprescu I, Morariu S, Ilie M, Hotea I, Imre M, Indre D, Balint A, Sorescu D (2010) Intraspecific characterization of some Cryptosporidium parvum isolates from calves and lambs in Western Romania using molecular techniques. Sci Parasitol 11:47–50
Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B (2013) Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect 141:1009–1020
Kaupke A, Kwit E, Chalmers RM, Michalski MM, Rzeżutka A (2014) An outbreak of massive mortality among farm rabbits associated with Cryptosporidium infection. Res Vet Sci 97:85–87
Lassen B, Ståhl M, Enemark HL (2014) Cryptosporidiosis—an occupational risk and a disregarded disease in Estonia. Acta Vet Scand 5:56–36
Leoni F, Amar C, Nichols G, Pedraza-Díaz S, McLauchlin J (2006) Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 55:703–707
Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, Pan W, Hu Y, Cao J (2014) Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis 14:25. doi:10.1186/1471-2334-14-25
Majewska AC, Werner A, Sulima P, Luty T (1999) Survey on equine cryptosporidiosis in Poland and the possibility of zoonotic transmission. Ann Agric Environ Med 6:161–165
Majewska AC, Werner A, Sulima P, Luty T (2000) Prevalence of Cryptosporidium in sheep and goats bred on five farms in west-central region of Poland. Vet Parasitol 89:269–275
Majewska AC, Werner A, Sulima P (2001) Występowanie kryptosporidiozy u bydła hodowlanego w jednym gospodarstwie rolnym—całoroczne badania. Wiad Parazytol 47(suppl 2):31
Majewska AC, Solarczyk P, Tamang L, Graczyk TK (2004) Equine Cryptosporidium parvum infections in western Poland. Parasitol Res 93:274–278
Millar C, Moore J, Lowery C, McCorry K, Dooley J (2001) Successful PCR amplification of genomic DNA from Cryptosporidium parvum oocysts extracted from a human faecal sample: a rapid and simple method suited for outbreak analysis. Int J Hyg Environ Health 204 :191 –194
Mišic Z, Abe N (2007) Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 134:351–358
Muhid A, Robertson I, Ng J, Ryan U (2011) Prevalence of and management factors contributing to Cryptosporidium sp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp Parasitol 127:534–538
O’Brien E, McInnes L, Ryan U (2008) Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp Parasitol 118:118–121
Olson ME, O’Handley RM, Ralston BJ, McAllister TA, Thompson RC (2004) Update on Cryptosporidium and Giardia infections in cattle. Trends Parasitol 20:185–191
Plutzer J, Karanis P (2007) Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet Parasitol 146:357–362
Quilez J, Torres E, Chalmers RM, Robinson G, Del Cacho E, Sanchez-Acedo C (2008) Cryptosporidium species and subtype analysis from dairy calves in Spain. Parasitology 135:1613–1620
Rieux A, Chartier C, Pors I, Delafosse A, Paraud C (2013) Molecular characterization of Cryptosporidium isolates from high-excreting young dairy calves in dairy cattle herds in Western France. Parasitol Res 112:3423–3431
Ryan U, Hijjawi N (2015) New developments in Cryptosporidium research. Int J Parasitol 45:367–373
Rzeżutka A, Kaupke A (2013) Occurrence and molecular identification of Cryptosporidium species isolated from cattle in Poland. Vet Parasitol 196:301–306
Rzeżutka A, Kaupke A, Kozyra I, Pejsak Z (2014) Molecular studies on pig cryptosporidiosis in Poland. Pol J Vet Sci 17:577–582
Santín M, Trout JM (2008) Livestock. In: Fayer R, Xiao L (eds) Cryptosporidium and cryptosporidiosis. CRC, Boca Raton, pp 451–483
Silverlås C, Näslund K, Björkman C, Mattsson JG (2010) Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol 169:289–295
Silverlås C, Bosaeus-Reineck H, Näslund K, Björkman C (2013) Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int J Parasitol 43:155–161
Soba B, Logar J (2008) Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology 135:1263–1270
Stantič-Pavlinic M, Xiao L, Glaberman S, Lal AA, Orazen T, Rataj-Verglez A, Logar J, Berce I (2003) Cryptosporidiosis associated with animal contacts. Wien Klin Wochenschr 115:125–127
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L (2005) Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43:2805–2809
Tanriverdi S, Arslan MO, Akiyoshi DE, Tzipori S, Widmer G (2003) Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol Biochem Parasitol 130:13–22
Thompson HP, Dooley JS, Kenny J, McCoy M, Lowery CJ, Moore JE, Xiao L (2007) Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol Res 100:619–624
Waldron LS, Ferrari BC, Power ML (2009) Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Exp Parasitol 122:124–127
Wang R, Zhang L, Axén C, Bjorkman C, Jian F, Amer S, Liu A, Feng Y, Li G, Lv C, Zhao Z, Qi M, Dong H, Wang H, Ning C, Sun Y, Xiao L (2014) Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Sci Rep 27:4208
Wielinga PR, de Vries A, van der Goot TH, Mank T, Mars MH, Kortbeek LM, van der Giessen JW (2008) Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int J Parasitol 38:809–817
Xiao L (2010) Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65:1578–1583
Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, Ling H, Liu A, Shen Y (2013) Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China’s Heilongjiang Province. PLoS ONE 8(1), e54857
Zintl A, Neville D, Maguire D, Fanning S, Mulcahy G, Smith HV, De Waal T (2007) Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitol 134:1575–1582
Acknowledgments
The authors thank Dr. Rachel M. Chalmers, Cryptosporidium Reference Unit in Swansea, UK, for her valuable comments on the manuscript; Prof. Mirosław M. Michalski, Department of Parasitology and Invasive Diseases, Faculty of Veterinary Medicine, University of Warmia and Mazury in Olsztyn, Poland, for the help in sampling arrangement; as well as Dr. Iwona Kozyra, Department of Food and Environmental Virology, National Veterinary Research Institute in Puławy, Poland, for the assistance in the graphical presentation of data. The study was supported by the Ministry of Science and Higher Education of Poland (research project no. S/071).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kaupke, A., Rzeżutka, A. Emergence of novel subtypes of Cryptosporidium parvum in calves in Poland. Parasitol Res 114, 4709–4716 (2015). https://doi.org/10.1007/s00436-015-4719-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-015-4719-1