Abstract
The presence of Cryptosporidium parasites in mammals and reptiles kept at the Lisbon Zoo was investigated. A total of 274 stool samples were collected from 100 mammals and 29 reptiles. The species and genotype of the isolates identified by light microscopy were determined by nested PCR and sequence analysis of a fragment of the small subunit rRNA gene. Cryptosporidium oocysts were found in one black wildebeest (Connochaetes gnou), one Prairie bison (Bison bison bison) and in one Indian star tortoise (Geochelone elegans). The PCR and sequence analysis of these three isolates showed that those excreted by the Prairie bison were Cryptosporidium mouse genotype, those from the black wildebeest were from a new Cryptosporidium genotype and those infecting the Indian star tortoise were Cryptosporidium tortoise genotype. The present work reports a new Cryptosporidium genotype in a black wildebeest and the first finding of the Cryptosporidium mouse genotype in a ruminant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium spp. are widespread protozoa considered to be important causes of gastrointestinal disease in humans and vertebrates.
Cryptosporidium infections have been reported in a wide spectrum of mammals belonging to several orders – Primata, Artiodactyla, Perissodactyla, Carnivora, Lagomorpha, Rodentia, Marsupialia, Monotrema and Proboscidea – and in numerous reptilian species belonging to orders Testudines and Squamata (Graczyk et al. 1997; Majewska et al. 1997; Mtambo et al. 1997; Muriuki et al. 1997; Gómez et al. 2000; Graczyk and Cranfield 2000). Recent molecular studies have shown that there is an extensive genetic diversity in Cryptosporidium parasites infecting mammals, reptiles and birds and in addition to the 14 Cryptosporidium accepted species – C. andersoni, C. hominis, C. parvum, C. canis, C. felis, C. wrairi, C. suis, C. muris, C. saurophilum, C. serpentis, C. baileyi, C. meleagridis, C. galli and C. molnari – many host-adapted genotypes have been described such as the deer, mouse, pig, bear, cervine, mongoose, marsupial, snake and lizard genotypes (Morgan et al. 1999a; Ryan et al. 2004; Xiao and Ryan 2004; Xiao et al. 2004a, 2004b).
Studies carried out previously in artiodactyl ruminants at the Lisbon Zoo, showed a 4% prevalence of cryptosporidiosis (Delgado et al. 2003) and molecular characterization has identified the isolates as C. parvum (Alves et al. 2001, 2003). In the present study, we extended the range of animals from the Lisbon Zoo examined for Cryptosporidium to several other mammalian orders and to reptiles. We also characterized part of the small subunit (SSU) rRNA gene of Cryptosporidium-positive samples by PCR amplification and sequence analysis.
Materials and methods
Between April 2002 and February 2003, 274 stool samples were collected from 100 mammals (Table 1) and 28 reptiles (Table 2) kept at the Lisbon Zoo.
With the exception of two baby lions, all mammals were adults, and had no Cryptosporidium-related symptoms. The reptiles studied, adults and young animals, were all asymptomatic, with the exception of one Indian star tortoise, which was part of a large group of confiscated tortoises in the black market in Singapore, that was severely sick due to crowded facilities, bad nutrition status, extreme temperature changes, and transport-induced stress. This tortoise arrived at the Lisbon Zoo 3 days prior stool collection and died 2 weeks later.
Direct and concentrated faecal smears were prepared and Cryptosporidium oocysts identified by light microscopy at a ×400 magnification after modified Ziehl-Neelsen staining. In samples that were positive by microscopy, oocysts’ DNA was extracted by a KOH/QIAamp DNA stool mini kit protocol (QIAGEN GmbH) (Alves et al. 2003). The species and genotype of the isolates identified were determined by nested PCR of a fragment of the small subunit (SSU) rRNA gene (Xiao et al. 2001) and sequencing of the secondary PCR products in both directions on an ABI Prism 3100 analyser (Applied Biosystems). The nucleotide sequences obtained were aligned with additional Cryptosporidium SSU rRNA sequences obtained from GenBank using the ClustalX program. A neighbour-joining tree was constructed from the aligned sequences with the TreeconW program using a sequence of Eimeria tenella (AF026388) as an outgroup. Genetic distance was calculated based on the Kimura 2-parameter model. Bootstrap analyses were conducted using 1,000 pseudo replicates.
Results
Oocysts were found in faeces of two artiodactyl bovids, a black wildebeest (Connochaetes gnou) and a Prairie bison (Bison bison bison), and in one Testudinidae, the Indian star tortoise (Geochelone elegans) that came from Singapore. In both mammals, the level of oocyst shedding was low and none had apparent symptoms of cryptosporidiosis. Subsequent stool collection was conducted on these two animals, but no oocysts were found. The level of oocyst shedding in the Indian star tortoise was low.
A fragment of the SSU rRNA gene was successfully amplified and sequenced for these three isolates. BLASTn GenBank searches performed on the sequences obtained from the Prairie bison and the Indian star tortoise, showed 100% similarity with the sequences of Cryptosporidium mouse genotype (AF112571) and Cryptosporidium tortoise genotype (AY120914), respectively (Xiao et al. 1999, 2004b).
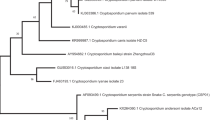
The black wildebeest isolate revealed a unique sequence, different from all other animal Cryptosporidium sequences, and thus constituted a new Cryptosporidium genotype in animals. The SSU rRNA partial nucleotide sequence of the black wildebeest Cryptosporidium isolate was deposited in the GenBank database under accession number AY883022. The new genotype from black wildebeest had the highest sequence homology to the W5 genotype (AF262332) previously found in a storm water sample collected in the Eastern Catskill Mountains, NY, with an A to G mutation, a TA insertion and a T deletion (Xiao et al. 2000). A neighbour-joining analysis showed that this genotype from wildebeest formed a cluster with several intestinal Cryptosporidium species (C. parvum, C. hominis, C. wrairi, C. suis, C. canis, C. felis, C. meleagridis and C. saurophilum) and genotypes (rabbit, horse, ferret, mouse, skunk, cervine, fox, squirrel, deer mouse, bear, opossum I and II, marsupial I and II, and muskrat I and II), with 87% of bootstrap support (Fig. 1).
Discussion
The present finding of Cryptosporidium mouse genotype in a Prairie bison constitutes the first report of this genotype in a ruminant. Gómez et al. (2000) also identified Cryptosporidium oocysts in a Prairie bison at the Barcelona Zoo, but the molecular identification was not performed. The mouse genotype is very common in mice and small rodents (Morgan et al. 1999b; Bajer et al. 2003). Whether the bison was infected with mouse genotype parasites or merely passing oocysts through the gastrointestinal tract after ingestion of food/water in facilities contaminated with rodents’ faeces remains to be determined. Phylogenetically, the mouse genotype has the closest relatedness to C parvum, which infects mainly ruminants (Xiao et al. 2004a). Thus, it is possible that the bison was actually infected by the mouse genotype. However, the absence of clinical signs, the low level of oocyst shedding and its detection only once in only one individual animal cannot exclude the possibility of the mere passage of oocysts in the animal. The turtle genotype found in the Indian star tortoise has already been found in three Indian star tortoises from the Saint Louis Zoo and thus should represent a true parasite of these reptiles (Xiao et al. 2004b).
Cryptosporidium infection is apparently common in wildebeests, as an earlier study identified Cryptosporidium spp. in 27% of black wildebeests from the Mikumi National Park, Morogoro, Tanzania (Mtambo et al. 1997). Because no molecular characterization of the isolates was done, the identity of Cryptosporidium spp. in these animals was not clear. Cryptosporidium was also previously found in faeces of a blue wildebeest (Connochaetes taurinus taurinus) at the Barcelona Zoo (Gómez et al. 2000). Molecular characterization of the parasite identified it as C parvum (Morgan et al. 1999b). The black wildebeest in this study was apparently passing oocysts of a new Cryptosporidium genotype. Due to its similarity with the W5 genotype from storm water, this new genotype may represent a new Cryptosporidium parasite that the black wildebeest acquired from other animals in the Zoo, rather than a native parasite from Africa. Thus, like many species or groups of vertebrates (Xiao et al. 2004a), wildebeests may be infected with at least two Cryptosporidium parasites, C. parvum and this new Cryptosporidium genotype.
References
Alves M, Matos O, Fonseca IP, Delgado E, Lourenço AM, Antunes F (2001) Multilocus genotyping of Cryptosporidium isolates from human HIV-infected and animal hosts. J Euk Microbiol (Suppl):17S-18S
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41:2744–2747
Bajer A, Cacciò S, Bednarska M, Behnke JM, Pieniazek NJ, Sinski E (2003) Preliminary molecular characterization of Cryptosporidium parvum isolates of wildlife rodents from Poland. J Parasitol 89:1053–1055
Delgado E, Fonseca IP, Fazendeiro I, Matos O, Antunes F, Cunha MB (2003) Cryptosporidium spp. in ruminants at the Lisbon Zoo. J Zoo Wildl Med 34:352–356
Gómez MS, Torres J, Gracenea M, Fernandez-Morán J, Gonzalez-Moreno O (2000) Further report on Cryptosporidium in Barcelona Zoo. Parasitol Res 86:318–323
Graczyk TK, Cranfield MR (2000) Cryptosporidium serpentis oocysts and microsporidian spores in feces of captive snakes. J Parasitol 86:413–414
Graczyk TK, Balazs GH, Work T, Aguirre AA, Ellis DM, Murakawa SKK, Morris R (1997) Cryptosporidium sp. infections in green turtles, Chelonia mydas, as a potential source of marine waterborne occysts in the Hawaiian Islands. Appl Environ Microbiol 63:2925–2927
Majewska AC, Kasprzak W, Werner A (1997) Prevalence of Cryptosporidium in mammals housed in Poznań Zoological Garden, Poland. Acta Parasitol 42:195–198
Morgan UM, Xiao L, Fayer R, Lal AA, Thompson RCA (1999a) Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int J Parasitol 29:1733–1751
Morgan UM, Strudee AP, Singleton G, Gómez MS, Gracenea M, Torres J, Hamilton SG, Woodside DP, Thompson RCA (1999b) The Cryptosporidium “mouse” genotype is conserved across geographic areas. J Clin Microbiol 37:1302–1305
Mtambo MMA, Sebatwale JB, Kambarage DM, Muhairwa AP, Maeda GE, Kusiluka LJM, Kazwala RR (1997) Prevalence of Cryptosporidium spp. oocysts in cattle and wildlife in Morogoro region, Tanzania. Prev Vet Med 31:185–190
Muriuki SMK, Farah IO, Kagwiria RM, Chai DC, Njamunge G, Suleman M, Olobo JO (1997) The presence of Cryptosporidium oocysts in stools of clinically diarrhoeic and normal nonhuman primates in Kenya. Vet Parasitol 72:141–147
Ryan UM, Monis P, Enemark HL, Sulaiman I, Samarasinghe B, Read C, Buddle R, Robertson I, Zhou L, Thompson RC, Xiao L (2004) Cryptosporidium suis n. sp. (Apicomplexa: Cryptosporidiidae) in pigs (Sus scrofa). J Parasitol 90:769–773
Xiao L, Ryan UM (2004) Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect Dis 17:483–490
Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RCA, Fayer R, Lal AA (1999) Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol 65:3386–3391
Xiao L, Alderisio K, Limor J, Royer M, Lal AA (2000) Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol 66:5492–5498
Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH, Lal AA (2001) Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis 183:492–497
Xiao L, Fayer R, Ryan U, Upton SJ (2004a) Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev 17:72–97
Xiao L, Ryan UM, Graczyk TK, Limor J, Li L, Kombert M, Junge R, Sulaiman IM, Zhou L, Arrowood MJ, Koudela B, Modrý D, Lal AA (2004b) Genetic diversity of Cryptosporidium spp. in captive reptiles. Appl Environ Microbiol 70:409–417
Acknowledgements
This work was supported by the projects POCTI/ESP/43635/99 and POCTI/ESP/46369/2002 from the Fundação para a Ciência e Tecnologia (FCT). Margarida Alves received a PhD thesis grant from FCT (SFRH/BD/2898/200). The authors would like to thank the workers of the Lisbon Zoo for their cooperation in sample collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Alves, M., Xiao, L., Lemos, V. et al. Occurrence and molecular characterization of Cryptosporidium spp. in mammals and reptiles at the Lisbon Zoo. Parasitol Res 97, 108–112 (2005). https://doi.org/10.1007/s00436-005-1384-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-005-1384-9