Abstract
Purpose
Abnormal expression of miRNAs is intimately related to a variety of human cancers. The purpose of this study is to confirm the expression of miR-181a and elucidate its physiological function and mechanism in pediatric acute myeloid leukemia (AML).
Methods
Pediatric AML patients and healthy controls were enrolled, and the expression of miR-181a and ataxia telangiectasia mutated (ATM) in tissues were examined using quantitative PCR. Moreover, cell proliferation and cell cycle were evaluated in several cell lines (HL60, NB4 and K562) by using flow cytometry after transfected with miR-181a mimics and inhibitors, or ATM siRNA and control siRNA. Finally, ATM as the potential target protein of miR-181a was examined.
Results
We found that miR-181a was significantly increased in pediatric AML, which showed an inverse association with ATM expression. Overexpressed miR-181a in cell lines significantly enhanced cell proliferation, as well as increased the ratio of S-phase cells by miR-181a mimics transfection in vitro. Luciferase activity of the reporter construct identified ATM as the direct molecular target of miR-181a. ATM siRNA transfection significantly enhanced cell proliferation and increased the ratio of S-phase cells in vitro.
Conclusion
The results revealed novel mechanism through which miR-181a regulates G1/S transition and cell proliferation in pediatric AML by regulating the tumor suppressor ATM, providing insights into the molecular mechanism in pediatric AML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) arises from the differentiation arrest of myeloid precursor and malignant proliferation in bone marrow and blood. It can be divided into different subtypes based on the cell type and the degree of maturity (Löwenberg et al. 1999). Although much progress has been made in the clinical treatment of AML in recent decades, only approximately 40 % of patients with AML achieved long-term survival (Estey and Döhner 2006). Therefore, a better understanding of the molecular mechanisms responsible for leukemogenesis and an exploration of novel therapeutic strategies are crucial for achieving an improved clinical outcome.
The biological processes of AML lineages is regulated by several lineage-specific regulators such as microRNAs (miRNAs), which have key roles in hematopoiesis (Chen 2005; O’Connell et al. 2011). miRNAs are non-coding RNAs of 19–25 nucleotides in length that regulate gene expression by repressing translation or accelerating mRNA decay (Bartel 2004). The dysregulated expression of specific miRNAs that modulates the expression of oncogenes or tumor suppressors is associated with the development and prognosis of malignancies (Esquela-Kerscher and Slack 2006; Huang et al. 2012; Yang et al. 2013; Cai et al. 2013; Ell et al. 2013). Recently, several studies have demonstrated that miR-181a is preferentially expressed in the B cells within mouse bone marrow, and its ectopic expression in hematopoietic stem/progenitor cells promotes B cell differentiation in both cultured tissues and peripheral blood of transplanted mice (Chen et al. 2004). miR-181a has been proposed to be involved in erythropoiesis (Choong et al. 2007). The expression changes of miR-181a, which are increased during T cell maturation (DN1–3) before dropping in CD4+ and CD8+ T cells, have also been shown to modulate TCR signaling (Li et al. 2007). miR-181a is capable of effectively repressing Lin28 expression, disrupting the Lin28–Let-7 reciprocal regulatory loop and eventually promoting MK differentiation (Li et al. 2012b). Moreover, miR-181a and miR-181b expression profoundly affected the development of human NK cells from CD34+ hematopoietic progenitor cells and IFN-γ production in primary CD56 + NK cells, suggesting it may play a critical role in the development of hematopoietic progenitor cells (Cichocki et al. 2011). However, the mechanism of miR-181a in regulating myeloid cells has not been fully understood.
We have previously shown that miR-181a is significantly upregulated in pediatric AML patients by microarray analysis (Zhang et al. 2009), which raises the possibility that miR-181a contributes to the development or pathogenesis of AML. However, the underlying molecular basis has not been elucidated. To address this issue, we examined the expression of miR-181a and investigated the potential molecular target of miR-181a in the pathogenesis of pediatric AML.
Materials and methods
Ethics statement
The research protocol was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University and Guangzhou Women and Children’s Medical Center. All participants with samples provided their written informed consent from the parents or guardians to participate in this study.
Subjects and samples
A total of 57 pediatric AML samples including five subtypes (from AML-M1 to AML-M5) from the First Affiliated Hospital of Sun Yat-sen University were enrolled in this study according to the following criteria: age ≤16 years at diagnosis,patients with primary AML and untreated. Patient characteristics were available (Supplement Table 1). Bone marrow was collected from patients by bone marrow puncture at the time of diagnosis.
As the healthy control, CD34+ cells isolated from 18 cord blood (CB) were included, which were obtained after normal full-term delivery of babies with the mothers’ informed consent of the biobank of Guangzhou Birth Cohort. CB was processed within 6 h of collection. Mononuclear cells were isolated by density gradient centrifugation at 400g for 30 min, washed twice with phosphate-buffered saline and resuspended in buffer solution. CD34+ cells were isolated by positive selection with an immunomagnetic separation system with CD34 MACS beads (Miltenyi Biotech, Gladbach, Germany). Purity of the sorted CD34+ cells was routinely >97 %. CD34+ hematopoietic progenitor cells and CD34− MNC fractions were cryopreserved and stored in −80 °C.
Cell lines and culture
Human leukemia cell lines NB4, HL60, MV-4-11, THP-1, K562, CCRF-CEM, Jurkat and SUP-B15 (all were purchased from American Type Culture Collection) were maintained in RPMI1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum (FBS). Human embryonic kidney cell line HEK-293T was maintained in DMEM medium supplemented with 10 % FBS. Cells were cultured at 37 °C with 5.0 % CO2 for further use.
DNA constructs
The pcDNA6.2-181a plasmid was generated by cloning a fragment containing the precursor of miR-181a-5p (from 1940 to 2153 bp) into the pcDNA6.2 vector (Invitrogen). The negative control plasmid, which contains an insert that can form a hairpin structure that can be processed into a mature miRNA, which is predicted not to target any known vertebrate gene, was named pcD6.2-NC. To construct psi-Check2–ATM–3′-UTR plasmid, a wild-type 3′-UTR segment of human ATM mRNA (3496–3547nt, NCBI Reference Sequence: NM_000051.3) containing the putative miR-181a-binding sequence (3525–3531nt) was synthesized and cloned into the XhoI and NotI sites downstream of the luciferase reporter gene in psi-Check2 vector (Promega, Madison, WI, USA).
This reporter construct was used as reported to determine the activity of miR-181a in inhibiting ATM expression through this site. As control, the empty psi-Check2 and a mutated reporter were used. The mutated reporter, psi-Check2–ATM–Mut, was generated by replacing the miR-181a targeting site with a mutated sequence. All cloned products were verified by sequencing. Primers and other oligonucleotide sequences were shown in Supplement Table 2.
Target gene prediction
The miRNA targets were analyzed and identified by three computer programs/algorithms, namely TARGETSCAN (http://www.targetscan.org), miRBase (http://www.mirbase.org/index.shtml) and PICTAR (http://pictar.mdc-berlin.de), which are commonly used in the field for the miRNA target prediction. To overcome the limitation of each program, the putative target genes predicted by at least two of the programs were selected for further experimental analyses.
Luciferase reporter assay
To determine the inhibitory effect of miR-181a on the expression of ATM, the luciferase reporter, carrying the 3′-UTR of ATM, its mutant sequence or pcDNA control vector, was cotransfected with vectors expressing miR-181a or control into HEK-293T cells for reporter assays. In brief, 200 ng of pcDNA–miR-NC/pcDNA–miR-181a and 50 ng of psi-Check2, psi-Check2–ATM–3′-UTR or psiCheck2–ATM–Mut were cotransfected into one well of HEK-293T cells (~5 × 104 cells), which were cultured on 48-well plate using Lipofectamine2000 (Life Technologies) according to the manufacturer’s instruction. Luciferase activity assay was performed at 48 and 72 h after transfection using Dual-Luciferase® Reporter 1000 Assay System (Promega). The effect of miR-181a on the expression of ATM was detected by the relative value of firefly luciferase activity normalized by Renilla luciferase activity. Each experiment was performed in triplicates.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from bone marrow samples using TRIzol® reagent (Life Technologies). Reverse transcription (RT) and qPCR were used to determine the expression level of miR-181a and ATM. In brief, RT was performed with 0.6 μg of total RNA for preparing cDNA using M-MLV reverse transcriptase (Promega), and then, qPCR was performed to amplify the specific target with specific mRNA/miRNA primers (Sangon, Shanghai, China) using The LightCycler® 480 Real-time PCR System (Roche, Basel, Switzerland) using SYBR® Premix Ex Taq™ II (TaKaRa, DaLian, China). The expression level was calculated using the \(2^{-\Delta \Delta {C_{\text{t}}}}\) method. The relative expression level of miRNA was obtained by the normalization to the values of U6. The relative expression level of ATM mRNA was obtained by the normalization to the values of GAPDH. The results were presented as the fold change of each target in the patient samples relative to the average value of that in healthy donor samples. All assays were performed in triplicates. The sequences for all primers used in the PCR were available (Supplement Table 2).
Cell electroporation
The miR-181a mimics (double strands) and scrambled oligonucleotides (referred to as miR-negative control; miR-NC) were purchased from GenePharma Biotech, Shanghai, China, which were used to evaluate miR-181a expression. The miR-181a antisense oligonucleotide (anti-miR-181a) and its counterpart control (anti-miR-NC), the small interfering RNAs (siRNAs) against human ATM (ATM–siRNA) and the negative control RNA duplex (NC–siRNA) were purchased from RiboBio, Guangzhou, China.
These small RNAs were delivered into cultured cells by electroporation. In brief, NB4, HL60 and K562 cells (2 × 105 cells/sample) were transfected with 100 pmol of the small RNAs using the Neon® Transfection System (Invitrogen). The electroporation conditions for these cells were optimized as follows: 1400 V, 10 ms width and 3 pulses for NB4 cells, 1350 V, 30 ms width and 1 pulse for HL60 cells, and 1450 V, 10 ms width and 3 pulses for K562 cells. The transfected cells were collected for cell proliferation and cell cycle assays as well as RNA and protein extraction for further analyses at different time points after electroporation.
Cell proliferation
NB4, HL60 and k562 cells were transfected with small RNAs by electroporation as described above. Cells were seeded into 96-well plates (2 × 103 cells/well) with 100 μL medium after electroporation. The relative amounts of viable cells were determined using a Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan) at each 24 h after electroporation till day 4. Firstly, 10 µL of CCK-8 solution was added to each well of the plate, then the plate was incubated for 2 h in the incubator, and the absorbance measured at 450 and 630 nm using a microplate reader. There were three wells in each treatment group at each time point, and the transfected small molecules were mentioned above. The experiment has been repeated for three times.
Cell cycle assay
NB4, HL60 and K562 cells that transfected with small RNAs were seeded into 12-well plate. After transfection for 48 h, cells were cultured in serum-free medium for 22 h and then FBS was supplied for additional 2 h. Then, the cells were harvested and washed with cold PBS twice, followed by incubation with NP40/propidium iodide (PI) dye solution containing RNaseA (30 mg/mL) for 30 min at 37 °C. After PI staining, cells were analyzed for the DNA content by FACS calibers (BD, NJ, USA) with CELL Quest software (Becton–Dickinson, San Jose, CA, USA). PI-negative living cells were gated for the cell cycle analysis. Each group was performed in triplicates.
Western blot analysis
Cells were collected in RIPA lysis buffer with protease and phosphatase inhibitors for 48 h after transfection. Lysates were incubated at 4–8 °C for 30 min, followed by centrifugation at 15,700g at 4–8 °C for 30 min. Meanwhile, the concentration was measured by BCA Protein Assay Kit (Pierce). Western blot was performed as follows: equal amounts of proteins (2 mg) and 10 mL loading control were separated by 6 % SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membranes were incubated with antibodies against ATM (Santa Cruz, Santa Cruz Biotech, CA, USA), then antibody binding was assessed by incubation with horseradish peroxidase-conjugated anti-mouse secondary antibodies (Santa Cruz). And the loading was monitored by β-tubulin or β-actin levels determined by anti-β-tubulin antibody or anti-β-actin antibody (Cell Signaling, Boston, MA, USA). At last, the protein level was detected using a luminal reagent (Millipore).
Statistical analysis
The Mann–Whitney U test was used to compare baseline characteristics and average miRNA/mRNA expression levels between patients and healthy individuals. The data of cytological experiments were expressed as the mean ± SD in at least three independent experiments. Unless otherwise noted, the differences between groups were analyzed using Student’s t test when there were only two groups, or assessed by one-way analysis of variance (ANOVA) when more than two groups were analyzed. Correlations were determined using the spearman correlation analysis. Analysis of term relapse-free survival (RFS)—defined as the time from complete remission to relapse—was performed according to the Kaplan–Meier method, and comparisons of outcomes between subgroups were performed by using the log-rank test. Differences were considered statistically significant at p < 0.05.
Results
The level of miR-181a is significantly upregulated in patients with primary pediatric AML
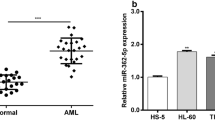
It is reported that miR-181a was upregulated in pediatric AML patients compared with normal control using microarray analysis performed previously by our group. To further confirm and extend this finding, in this study, we examined miR-181a expression by qPCR in cell lines, pediatric AML patient samples and normal CB CD34+ cells, respectively. As shown in Fig. 1, higher expression level of miR-181a was detected in 3 AML cell lines (HL60, NB4 and MV-4-11) but not THP-1 and K562 when compared with CB CD34+ cells (p < 0.01). Also, miR-181a was higher in some acute lymphoblastic leukemia (ALL) cell lines (CCRF-CEM). Moreover, miR-181a expression was significantly higher in pediatric AML samples compared to healthy control (p < 0.05). Of them, 35 of 57 (61.4 %) AML displayed more than twofold upregulation in the miR-181a expression level especially. Further, there was no significant difference in patient age, gender, white blood cell (WBC) count and some fusion genes between patients with low and high miR-181a expression except in morphological sub-class of pediatric AML when the relationship between miR-181a expression and clinicopathologic characteristics was analyzed (p = 0.03, Table 1).
The expression of miR-181a was dysregulated in myeloid cell lines and clinical specimens. a Expression of miR-181a in leukemia cell lines and hematopoietic cells. The fold induction expression is relative to CB CD34+ cells. b The average expression level of miR-181a in pediatric acute myeloid leukemia (AML) specimens (n = 57) and control (n = 18). The level of miRNA was normalized to the expression of U6, as quantified by real-time PCR. **p < 0.01; *p < 0.05. M1 AML without differentiation, M2 AML with differentiation, M3 acute promyelocytic leukemia, M4 acute myelomonocytic leukemia and M5 acute monoblastic leukemia
Overexpression of miR-181a promotes G1/S transition and cell proliferation in AML cells
To validate the aforementioned possible role of miR-181a in the pathogenesis of pediatric AML, we evaluated cell proliferation and cell cycle progression in NB4 cells in response to miR-181a transfection. We observed significantly time-dependently increased miR-181a expression after miR-181a mimics transfection compared with the control group (p < 0.01). Similar upregulation of miR-181a expression was observed in HL60 and K562 cell lines (data not shown). Next, by using the CCK-8 assay, we examined cell proliferation in three cell lines (NB4, HL60 and K562) in response to miR-181a transfection. As shown in Fig. 2a–c, we found transfection of miR-181a mimics significantly increased cell proliferation in three cell lines (NB4, HL60 and K562) compared with normal control (p < 0.01). These findings suggested that miR-181a may contribute to AML progression by promoting myeloid cell proliferation.
Enforced expression of miR-181a induced cell proliferation of AML cells. a–c Effect of miR-181a on cell proliferation was measured by CCK-8 kit after miRNAs was transfected into NB4, HL60 and K562, respectively. d, e The cell cycle distribution of NB4, which was transfected with miRNAs for 48 h and then cultured in non-serum mediums for 22 h, and cell cycle phase distribution was analyzed by flow cytometry after PI staining. The percentage of cells in each phase is indicated. Representative data from three independent experiments are shown. f, g The cell cycle distribution of K562; the treatment was similar to previous. The data corresponded to the mean of three independent experiments. *p < 0.05, **p < 0.01
To assess whether miR-181a participated in the regulation of cell cycle, we examined G1/S transition in two cell lines (NB4 and K562) after miR-181a transfection by adopting a serum starvation–stimulation assay. NB4 and K562 cells were transfected with miR-181a mimics or control dsRNA for 48 h, followed by synchronization with serum deprivation for 24 h. Next, they were stimulated to enter S-phase by serum re-addition before flow cytometry analysis. As shown in Fig. 2d–g, we found significantly decreased ratio of G1-phase cells and significantly increased ratio of S-phase cells in miR-181a-transfected cells compared to the control group (p < 0.05). These findings indicated that overexpression of miR-181a contribute to cell cycle progression.
ATM is a direct molecular target of miR-181a
We predicted the possible target molecules of miR-181a by applying several miRNA target gene prediction websites and identified ATM as the candidate gene. As shown in Fig. 3b, seven nucleotides at the 5′end of miR-181a were perfectly complementary to the target sequence in the 3′-UTR of ATM (including the “seed sequence” from positions 2–8), and the predicted miR-181a-binding 3′-UTR of ATM is conserved in humans, Chimpanzee and Rhesus. To validate this, we cloned the ATM 3′-UTR portion containing the miR-181a target site into a Renilla luciferase reporter construct (Fig. 3b) and established a luciferase reporter assay following co-transfection of reporter constructs with precursor miR-181a-5p (pre-miR-181a) into HEK-293T cells. Consequently, a significant reduction in the luciferase activity of the reporter construct containing the ATM 3′-UTR was observed in the presence of miR-181a (55 %, p < 0.01). Full mutation of putative miR-181a-binding sites in the 3′-UTR of the ATM gene conversely rescues the reduced luciferase activity (Fig. 3c). To further validate this, we examined the expression of ATM in NB4 cells transfected with miR-181a mimics or inhibitors. The transfection efficiency, which was determined by qPCR, indicated miR-181a was significantly up- or downregulated in NB4 cells as expected (Fig. 3d). Accordingly, the level of ATM protein was significantly inhibited in miR-181a mimics-transfected NB4 cells, but significantly increased in inhibitor ones (Fig. 3e). Meanwhile, the same results were found in HL60 cells (Fig. 3f). These findings provide the evidence that ATM is a direct target of miR-181a in myelopoiesis.
ATM was a putative target gene of miR-181a. a Sequences of the predicted miR-181a-binding sites in human, Chimpanzee and Rhesus genomic regions. Highly conserved nucleotides are shown in bold. b Schematic of the luciferase reporter assay was used to validate the interaction between miR-181a and the 3′-UTR of ATM. Bold font indicates the “seed” regions. miRNA recognition elements of ATM 3′-UTR with wild and full mutant were separately inserted into a psiCheck-2 vector downstream from the Renilla luciferase gene. c Repression of luciferase activity due to the interaction between miR-181a and the luciferase constructs. Each Renilla luciferase reading was normalized to that obtained for the control firefly luciferase. d NB4 cells were electroporated with miR-NC, miR-181a, anti-miR-NC and anti-miR-181a, respectively. The levels of miR-181a were examined by qPCR. e NB4 cells lysates were prepared for western blot with an antibody against ATM after transfection, and the expression of β-tubulin severed as a loading control. f HL60 Cells lysates were prepared for western blot with an antibody against ATM after transfected, and the expression of β-actin severed as a loading control. **p < 0.01
ATM knock-down promoted cell proliferation and G1/S transition in human AML cells
Next, we assessed the possible role of ATM in cell proliferation and G1/S transition. As shown in Fig. 4, when compared with the control group, we found ATM siRNA transfection caused significant inhibition of ATM mRNA and protein expression in NB4 cells (p < 0.01), and significantly enhanced cell proliferation in both NB4 and K562 cells (p < 0.01, from 72 to 96 h). In the following studies, a serum starvation–stimulation assay was employed to evaluate whether ATM contributed to the cell cycle progression. NB4 and K562 cells were transfected with ATM siRNA or control siRNA for 48 h, followed by synchronization with serum deprivation for 22 h, respectively. As demonstrated by flow cytometry, we found significantly decreased ratio of G1-phase cells and significantly increased ratio of S-phase cells in ATM siRNA-transfected cells compared to the control group (p < 0.05). These findings indicated that inhibition of ATM contribute to cell cycle progression.
ATM was involved in miR-181a-induced cell proliferation in AML cells. a, b qRT-PCR and western blot analysis of the effects of ATM siRNA. NB4 cells were transfected with NC siRNA or ATM siRNA. c Effect of ATM siRNA on cell proliferation was measured by CCK-8 kit after siRNA transfected into NB4. d Effect of ATM siRNA on cell proliferation was measured by CCK-8 kit after siRNA transfected into K562. e, f The cell cycle distribution of NB4, which was transfected with siRNA for 48 h and then cultured in non-serum mediums for 22 h and cell cycle phase distribution was analyzed by flow cytometry after propidium iodide staining. Percentage of cells in each phase was indicated. Representative data from three independent experiments were shown. g, h The cell cycle distribution of K562; the treatment was similar to previous. The data corresponded to the mean of three independent experiments. *p < 0.05, **p < 0.01
Clinical relevance of miR-181a and ATM in pediatric primary AML
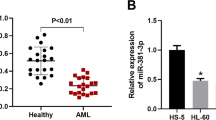
To assess the clinical relevance of miR-181a and ATM in pediatric AML, we examined the expression of miR-181a and ATM and evaluated the association between miR-181a and ATM in pediatric AML. The expression of ATM was detected in newly diagnosed pediatric AML, and 18 CB CD34+ samples from healthy donors were analyzed as controls. As shown in Fig. 5a, we found ATM is significantly downregulated in pediatric AML patients (p < 0.05). When correlating ATM with miR-181a expression in pediatric AML patients, we found a significantly inverse association of ATM and miR-181a expression in this cohort (r = −0.598, p < 0.001).
ATM was downregulated in pediatric AML specimens and inversely correlated with miR-181a levels. a The expression level of ATM in pediatric AML specimens and normal controls. ATM level was normalized to GAPDH. *p < 0.05. b A statistically significant inverse correlation between miR-181a and ATM mRNA level in pediatric AML specimens (Spearman’s correlation analysis, r = −0.598, p < 0.001)
In the following studies, 27 pediatric AML patients with complete follow-up data (Supplement Table 1; mean follow-up time was 88 months) were subdivided into two groups: those who developed distant relapse or death and those who were in continuous complete remission within the follow-up period. The expression level of miR-181a was slightly elevated in the primary AML of six patients with distant relapse or death, as compared with that in primary AML removed from 21 patients in complete remission, but without statistical significance (Supplement Figure 1a). The Kaplan–Meier survival plot was generated based on the miRNA of two groups, and no clear correlation between the level of miR-181a at diagnosis and RFS was identified (p = 0.4659) (Supplement Figure 1b).
Discussion
MiRNAs recently have emerged as important cellular regulator that govern cellular proliferation and differentiation, and dysregulation of cell cycle-related miRNAs may facilitate tumorigenesis (Kastan and Bartek 2004; Ventura and Jacks 2009). In this study, we showed that miR-181a was significantly upregulated in both bone marrow of pediatric AML patients and several AML cell lines. Moreover, our findings showed that overexpressed miR-181a promotes G1/S checkpoint transition and cell proliferation in vitro, which may expand our understanding on the pathophysiology of pediatric AML.
miR-181a has been proposed to be aberrantly expressed in a number of carcinomas, but its exact role in tumorigenesis is still controversial. Some studies have reported that miR-181 has been significantly upregulated in cancers of breast (Bisso et al. 2013; Taylor et al. 2013), liver (Wang et al. 2010) and blood (Pichiorri et al. 2008; Pons et al. 2009) as the promoting factor of carcinogenesis. In contrast, some studies proposed that miR-181a was significantly downregulated in solid tumors as tumor suppressor (Iliopoulos et al. 2010; Shin et al. 2011; Shi et al. 2008; Ouyang et al. 2012). In adult acute leukemia, increased expression levels of miR-181a have been shown to be associated with favorable outcome of cytogenetically normal AML patients; miR-181a overexpression thus is considered as a favorable prognostic marker in this setting (Marcucci et al. 2008; Schwind et al. 2010; Li et al. 2012c). However, Zhuang et al. (2014) recently reported that, in 62 patients with acute promyelocytic leukemia (21 newly diagnosed, 19 in complete remission and 22 relapsed) and 15 non-leukemia patients, a relatively higher expression of miR-181a was found in newly diagnosed or relapsed patients, but a very low expression in both non-leukemia patients and acute promyelocytic leukemia patients in complete remission, suggesting miR-181a may promote the development of acute promyelocytic leukemia.
Currently, the importance of miR-181 in pediatric AML has been well documented. We previously reported miR-181a was significantly increased in pediatric AML patients by microarray analysis (Zhang et al. 2009). Consistently, we observed miR-181a expression level was significantly increased in pediatric AML patients compared with healthy controls using qPCR analysis. Interestingly, when subclassifing the AML samples into five subtypes (M1–M5) based on the morphological FAB phenotype, we found miR-181a levels were significantly elevated in M1 or M2 or M3 subtypes compared with M4 or M5 morphology in pediatric samples, whose results were similar to that in adult AML (Debernardi et al. 2007).
It is known that miRNA can regulate a large fraction of protein-coding genes, which form a regulatory network during cell differentiation and development. For example, Wang et al. (2009) reported that miR-181 regulated the expression of p27Kip1 in human myeloid leukemia cells and induced the differentiation by 1,25-dihydro-xyvitamin D3 (1,25-D3). Recently, Zhai et al. (2013) showed that increased expression of miR-181a induced apoptosis by repressing its target gene BCL-2 in prostate cancer PC-3 cell line. To examine the direct molecular target of miR-181a in pediatric AML, we predicated the candidate gene by applying several miRNA target gene prediction websites and identified ATM as the candidate gene, which was further validated by luciferase activity assay. ATM is a key regulator of the DNA damage response through phosphorylating a variety of proteins involved in DNA repair, cell cycle regulation and apoptosis (Kastan and Lim 2000). In the present study, we provided the evidence that ATM expression was significantly inhibited, which was inversely associated with miR-181a expression in pediatric AML. This finding provides the clue that ATM may be the direct molecular target of miR-181a myeloid cells. Recently, Bisso et al. (2013) reported that miR-181a and miR-181b were overexpressed in more aggressive breast cancer, and their expression inversely correlates with ATM levels as well. However, the possible role of ATM in cell differentiation has not been well characterized. In this study, we reported that ATM knock-down promoted cell proliferation and G1/S transition in human AML cells, suggesting miR-181a and ATM may contribute to the pathogenesis of pediatric AML by regulating cell development and differentiation.
The clinical relevance of increased miR-181a and ATM expression in pediatric AML remains unclear. Although overexpression of miR-181a has been proposed as a favorable prognostic marker in adult acute leukemia, we only found the expression level of miR-181a was slightly elevated in the primary AML of six patients with distant relapse or death, as compared with that in primary AML removed from 21 patients in complete remission, but without statistical significance in the present study. The Kaplan–Meier survival plot was generated based on the two groups of miRNAs, and no clear correlation between the levels of miR-181a at diagnosis and RFS as identified. The lack of statistical significance may be ascribed to the limited samples as well. Recently, several studies have also demonstrated that miR-181a sensitizes K562 and HL60 to daunorubicin and Ara-C, respectively (Bai et al. 2012; Li et al. 2012a). Since overexpression of miR-181a has been found to promote G1/S transition of AML cells, it may be interesting to perform further study to clarify whether miR-181a can make AML cell lines more sensitive to chemotherapy in the clinical settings.
Conclusion
We provided the evidence that miR-181a was significantly upregulated in pediatric AML with respect to healthy controls, which showed an inverse association with ATM expression. Moreover, we identified ATM as the direct molecular target of miR-181a by luciferase activity of the reporter construct and showed that overexpression of miR-181a in cell lines significantly enhanced cell proliferation and increased the ratio of S-phase cells by suppressing ATM expression in vitro. Our results provided insights into the molecular mechanism underlying pediatric AML.
References
Bai H, Cao Z, Deng C, Zhou L, Wang C (2012) miR-181a sensitizes resistant leukaemia HL-60/Ara-C cells to Ara-C by inducing apoptosis. J Cancer Res Clin Oncol 138(4):595–602
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bisso A, Faleschini M, Zampa F, Capaci V, De Santa J et al (2013) Oncogenic miR-181a-5p/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 12:1679–1687
Cai J, Guan H, Fang L, Yang Y, Zhu X et al (2013) MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest 123:566–579
Chen CZ (2005) MicroRNAs as oncogenes and tumor suppressors. N Engl J Med 353:1768–1771
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86
Choong ML, Yang HH, McNiece I (2007) MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol 35:551–564
Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A et al (2011) Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol 187:6171–6175
Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-Mclver A et al (2007) MicroRNA miR-181a-5p correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 21(5):912–916
Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H et al (2013) Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 24:542–556
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Estey E, Döhner H (2006) Acute myeloid leukaemia. Lancet 368:1894–1907
Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES (2012) MiR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating FAS expression. Cancer Res 72:908–916
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K (2010) STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 39:493–506
Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432:316–323
Kastan MB, Lim DS (2000) The many substrates and functions of ATM. Nat Rev Mol Cell Biol 1:179–186
Li QJ, Chau J, Ebert PJ, Sylvester G, Min H et al (2007) miR-181a-5p is an intrinsic modulator of T cell sensitivity and selection. Cell 129:147–161
Li H, Hui L, Xu W (2012a) miR-181a sensitizes a multidrug-resistant leukemia cell line K562/A02 to daunorubicin by targeting BCL-2. Acta Biochim Biophys Sin (Shanghai) 44(3):269–277
Li X, Zhang J, Gao L, McClellan S, Finan MA et al (2012b) miR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ 19:378–386
Li Z, Huang H, Li Y, Jiang X, Chen P et al (2012c) Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood 119(10):2314–2324
Löwenberg B, Downing JR, Burnett A (1999) Acute myeloid leukemia. N Engl J Med 341:1051–1062
Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS et al (2008) MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 358(18):1919–1928
O’Connell RM, Zhao JL, Rao DS (2011) MicroRNA function in myeloid biology. Blood 118:2960–2969
Ouyang YB, Lu Y, Yue S, Giffard RG (2012) miR-181 targets multiple Bcl-2 family members and influences apoptosis and mitochondrial function in astrocytes. Mitochondrion 12:213–219
Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T et al (2008) MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA 105:12885–12890
Pons A, Nomdedeu B, Navarro A, Gaya A, Gel B et al (2009) Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma 50:1854–1859
Schwind S, Maharry K, Radmacher MD, Mrózek K, Holland KB et al (2010) Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol 28(36):5257–5264
Shi L, Cheng Z, Zhang J, Li R, Zhao P et al (2008) Hsa-miR-181a-5p and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res 1236:185–193
Shin KH, Bae SD, Hong HS, Kim RH, Kang MK et al (2011) miR-181a-5p shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem Biophys Res Commun 404:896–902
Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP (2013) TGF-β upregulates miR-181a-5p expression to promote breast cancer metastasis. J Clin Invest 123:150–163
Ventura A, Jacks T (2009) MicroRNAs and cancer: short RNAs go a long way. Cell 136:586–591
Wang X, Gocek E, Liu CG, Studzinski GP (2009) MicroRNAs181 regulate the expression of p27Kip1 in human myeloid leukemia cells induced to differentiate by 1,25-dihydroxyvitamin D3. Cell Cycle 8:736–741
Wang B, Hsu SH, Majumder S, Kutay H, Huang W et al (2010) TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29:1787–1797
Yang H, Cho ME, Li TW, Peng H, Ko KS et al (2013) MicroRNAs regulate methionine adenosyltransferase 1A expression in hepatocellular carcinoma. J Clin Invest 123:285–298
Zhai XF, Fang FF, Liu Q, Meng YB, Guo YY et al (2013) miR-181a-5p contributes to bufalin-induced apoptosis in PC-3 prostate cancer cells. BMC Complement Altern Med 13:325
Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS et al (2009) MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One 4(11):e7826
Zhuang LK, Xu GP, Pan XR, Lou YJ, Zou QP et al (2014) MicroRNA-181a-mediated downregulation of AC9 protein decreases intracellular cAMP level and inhibits ATRA-induced APL cell differentiation. Cell Death Dis 5(4):e1161
Acknowledgments
This work was supported by National Nature and Science Grant of China (81272310).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaodan Liu, Wang Liao and Hongxia Peng authors have contributed equally to the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Fig. 1
The relationship of miR-181a expression and prognosis in 27 AML patients with complete follow-up data. a miR-181a expression in patients who achieved a complete remission versus patients who developed distant relapse or death. b Relapse-free survival according to miR-181a expression levels in pediatric AML patients with high (above the median miR-181a expression value) or low (at or below the median miR-181a expression value) expression groups. Statistical differences between curves were calculated using the log-rank test (TIFF 10822 kb)
Supplement Table 1
Characteristics of pediatric AML patients (N = 57) (DOCX 17 kb)
Supplement Table 2
Primers and other oligonucleotides sequences (DOCX 14 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Liao, W., Peng, H. et al. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol 142, 77–87 (2016). https://doi.org/10.1007/s00432-015-1995-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-015-1995-1