Abstract
MicroRNA (miR)-381-3p is the newly discovered tumor-associated miRNA, which is frequently associated with diverse human malignancies; but, it is still unknown about its effect on acute myeloid leukemia (AML) in children. This work focused on exploring miR-381-3p’s effect on childhood AML and identifying the possible mechanisms facilitating new treatment development. Using qRT-PCR analysis, miR-381-3p expression remarkably reduced in pediatric AML patients and AML cell lines (HL-60 and U937). Following transfection of miR-381-3p mimic or inhibitor into HL-60 and U937 cells, we conducted MTT assay to evaluate cell proliferation, flow cytometry (FCM) to measured cell apoptosis and cell cycle, whereas Transwell assays to detect cell invasion and migration. Our results demonstrated that miR-381-3p overexpression remarkably repressed cell growth, invasion and migration; additionally, miR-381-3p overexpression resulted in arrest of cell cycle and enhanced cell apoptosis. In contrast, miR-381-3p knockdown led to an opposite effect. Moreover, we predicted miR-381’s target gene and validated it by luciferase reporter assay and TargetScan, separately. We identified miR-381-3p’s binding site in ROCK1 3′-UTR. As revealed by Western-blot (WB) assay, miR-381-3p overexpression notably suppressed ROCK1 level. Moreover, restoring ROCK1 expression abolished miR-381-3p’s inhibition on cell proliferation, invasion and migration. Data in this work indicated the role of miR-381-3p as the tumor suppressor within pediatric AML by targeting ROCK1. Therefore, miR-381-3p might serve as a potential therapeutic target for the treatment of pediatric AML.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myeloid leukemia (AML) represents the highly malignant hematopoietic neoplasm, which is associated with the features of disordered differentiation of healthy hematopoietic cells, together with ectopic immature myeloid cell accumulation (Oke et al. 2009; Alcalay et al. 2005). It is reported that overall survival (OS) rate of children with AML has currently reached 70% in recent clinical trials (Zwaan et al. 2015; Rubnitz et al. 2019; Pession et al. 2013), even though standard chemotherapy regimens have been intensified (Bachas et al. 2010; Brown et al. 2004). Thus, novel and effective therapeutic strategies are urgently needed. Molecular targeting therapy may be a promising candidate treatment for pediatric AML.
MicroRNAs (miRNAs) are single-stranded, small (~ 21 nucleotides), non-coding RNAs (ncRNAs), which play their functions in combination with target mRAN 3′-untranslated regions (3′-UTR) (Zheng et al. 2011). MiRNAs are associated with different pathological and biological events, such as cell proliferation, apoptosis, differentiation, migration, and invasion (Hu et al. 2014; Li et al. 2013). Numerous studies have demonstrated that the dysregulation of miRNA expression plays a critical role in human tumor occurrence and progression. MiR-381-3p, a novel cancer-related miRNA, is related to several human cancers, such as hepatocellular cancer (HCC) (Zhang et al. 2016), breast cancer (BC) (Shalini et al. 2016), osteosarcoma (Wang et al. 2013), gastric cancer (GC) (Zhang et al. 2017), and oral squamous cell carcinoma (OSCC) (Yang et al. 2017). As the serine/threonine kinase, Rho-associated-coiled coil-containing kinase 1 (ROCK1) is involved in multiple biological processes, containing actin cytoskeleton organization, cell adhesion, and mobility (Cai et al. 2015; Akagi et al. 2014). Previous studies have found that activating ROCK1 via Rho GTPases enhances cell migration as well as invasion of prostate cancer (PCa) (Kroiss et al. 2015), bladder cancer (Wu et al. 2016), BC (Raviraj et al. 2012), and pancreatic cancer (Kaneko et al. 2002). ROCK-LIM kinase (LIMK)-Cofilin axis plays an important role in regulating cytoskeleton assembly (Wang et al. 2019, 2018) and leukemic aggressiveness (Zampini et al. 2018; Djamai et al. 2021). A recent study showed that miR-381 was downregulated in bone marrow tissues and serum of pediatric AML patients and its serum level was an independent predictor of prognosis (Zhang et al. 2020). However, the role of miR-381 in pediatric AML still needs further elucidation.
This work aimed to examine miR-381-3p’s effect on pediatric AML, and to elucidate those possible mechanisms. As a result, miR-381-3p was obviously downregulated in bone marrow from children with AML. Moreover, our results showed that miR-381-3p overexpression suppressed cell growth, apoptosis and arrest of cell cycle, while suppressing cell invasion and migration through targeting ROCK1.
Materials and methods
Patients as well as tissue specimens
We obtained bone marrow specimens in 34 children with AML, and 21 healthy children as the normal controls, from January 2010 to December 2014 at Ningbo First Hospital of Zhejiang University (Ningbo, China). The clinical characteristics of 34 patients with AML are summarized in Table 1. Later, mononuclear cells were isolated from bone marrow samples using Ficoll-Paque™ PLUS (GE Healthcare Life Sciences, Freiburg, Germany) and preserved under − 80 °C for further studies. Our experiments were conducted following the 1975 Declaration of Helsinki guidelines and gained approval from Medical Ethics Committee of Ningbo First Hospital of Zhejiang University. Informed consent was obtained from each patient.
RNA extraction together with qRT-PCR
We utilized RNeasy Mini kit (Qiagen, Hilden, Germany) in order to extract total miRNA and RNA following specific instructions. Thereafter, total RNA content was quantified by Nanodrop 2000 (Thermo Fisher Scientific, Waltham, USA), followed by synthesis of cDNA with total RNA extracted by TaqMan High-Capacity cDNA Archive kit (Applied Biosystems, Foster, USA) through reverse transcription. To analyze mRNA level, TaqMan Universal PCR Master Mix (Applied Biosystems) was blended with reverse-transcribed products, followed by qRT-PCR carried out using the Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems). Sequences of specific primers (Invitrogen, Shanghai, China) were showed below, ROCK1, 5′-GGTGGTCGGTTGGGGTATTTT-3′ (F) and 5′-CGCCCTAACCTCACTTCCC-3′ (R); GAPDH, 5′-ACAACTTTGGTATCGTGGAAGG-3’ (F), and 5′-GCCATCACGCCACAGTTTC-3′ (R), with GAPDH being the internal reference for normalizing ROCK1 expression.
To detect miRNA level, we used miRNeasy Mini kit (Qiagen, Hilden, Germany) to extract miRNAs and adopted TaqMan MicroRNA Assay Kit for detecting miRNA levels based on the Applied Biosystems 7500 Fast Real-Time PCR systems. The universal small nuclear RNA, U6, served as the endogenous reference. 2-∆∆Ct approach was utilized to determine relative gene levels.
Cell culture and transfection
We obtained human HL-60 and U937 cells as well as the normal bone marrow stromal cell line HS-5 in Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China), and cultivated them within RPMI-1640 that contained 10% FBS (Gibco BRL, Rockville, USA) as well as 1% penicillin/streptomycin (P/S), followed by incubation under 37 °C and 5% CO2 conditions. We collected cells at exponential growth phase in further assays. In addition, cells were treated with 20 µM Y-27632 (Sigma-Aldrich, St. Louis, MO, USA) for 48 h at 37 °C with 5% CO2.
We adopted Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, USA) for cell transfection in line with specific protocols. Following transfection with miR-381-3p mimic/inhibitor, pcDNA3.1-ROCK1 or corresponding negative controls, designed by Shanghai GenePharma Co., Ltd. (Shanghai, China), HL-60 and U937 cells (5 × 105 cells/well) were cultivated into each well of 6-well plates until 70–80% confluency. After transfection for 2 days, we collected cells to conduct later assays.
Cell proliferation analysis (MTT assay)
We utilized the MTT Assay Kit (Sigma-Aldrich, USA) to assess cell proliferation in line with specific protocols. Following transfection, HL-60 and U937 cells were transferred to a 96-well plate. At the indicated time point (0 h, 12 h, 24 h, 48 h, and 72 h), MTT solution (20 μL) was placed into every well and incubated for another 4 h. The medium was discarded and 150 μL dimethyl sulfoxide (DMSO) per well was added to dissolve the formosan. A microplate reader (Bio-Tek Instruments, Germany) was used to examine the absorbance values at 490 nm.
Cell cycle and apoptosis assays
In order to analyze cell cycle distribution, we collected cells after transfection for 48 h. After rinsing by PBS, we fixed cells with ethanol under − 20 °C, rinsed them by PBS again, followed by rehydration and PI staining (BD Biosciences, San Jose, USA). We later adopted the flow cytometer (BD Biosciences) to analyze stained cells (~ 1 × 105), and examine them using ModFit software (BD Biosciences, San Jose, USA).
To analyze cell apoptosis, we collected cells and rinsed them by pre-chilled PBS, followed by staining using the Annexin V-fluorescein isothiocyanate apoptosis detection kits (BD Biosciences). We later adopted flow cytometer to measure stained cells. Early (Annexin V+/PI−)- and late (Annexin V+/PI+)-phase apoptotic cells were segregated with a quadri-plot graph, and the apoptosis was scored by quantifying the population of Annexin V-positive cells.
Cell migration and invasion assays
We conducted Transwell assay to measure cell migration using the chamber with 8-μm pore filters (Sigma-Aldrich). Briefly, after cell transfection, we inoculated cells (2 × 104) on top chamber that contained serum-free medium (200 μL). Meanwhile, we added a 750-μL medium that contained 10% FBS into the bottom chamber to incubate under 37 °C for a 24-h period. Afterwards, cells were subject to 4% paraformaldehyde (PFA) as well as staining using 0.5% crystal violet (Sigma-Aldrich). We later determined the stained cell number microscopically and chose 5 fields of view (FOVs) to determine the mean value. For invasion assay, we coated Matrigel (50 μL, 1:2; BD Biosciences, Franklin Lakes, USA) into the upper chamber. The rest of invasion assay was performed as migration assay.
Bioinformatic analysis as well as luciferase reporter assay
This work adopted the TargetScan online software for predicting miR-381-3p’s candidate target genes. Among all those genes that were compiled, we determined the genes that might contribute to pediatric AML progression, and analyzed the putative binding sequences.
We amplified 3′ UTR in ROCK1 that contained miR-381-3p’s-binding site in the human cDNA library. Afterwards, it was inserted in the dual-luciferase expression vector (Promega, Madison, WI, USA). Moreover, we acquired the mutant ROCK1 3′-UTR through overlap-extension PCR. Using Lipofectamine 2000, we co-transfected ROCK1 MUT or WT 3′-UTR with miR-381-3p mimics into HL-60 and U937 cells. Later, dual-luciferase reporter assay system (Promega Corp., Fitchburg, USA) was adopted to measure relative luciferase activity after transfection for 48 h. Results were presented in a form of luciferase activity ratio of Renilla to firefly.
Western-blot (WB) analysis
This study utilized radioimmunoprecipitation buffer (500 μL) that contained phenylmethane sulfonyl fluoride (1 mM) to extract cellular protein lysates. Afterwards, each sample was subject to 2-min sonication and centrifugation to collect collecting supernatants in later assays. We later separated lysates through 8% SDS-PAGE, followed by transfer on PVDF membrane. Later, we adopted PBS that contained 5% skimmed milk (w/v) and 0.1% Tween-20 (PBST) to block membranes under ambient temperature for a 1-h period. Membranes were later rinsed by PBST, incubated using anti-GAPDH (1:3000) and anti-ROCK1 (1:1000) antibodies (Abcam, Cambridge, MA, USA) under 4 °C overnight. After rinsing by PBST again, we added the horseradish peroxidase (HRP)-conjugated IgG (1:5000) for another 1-h incubation under ambient temperature. ECL WB reagents were employed for blot developing.
Statistical analysis
Results were presented in a form of mean ± SD. We adopted SPSS18.0 (SPSS Inc., Chicago, IL, USA) in performing statistical analyses. Differences of 2 groups were compared by carrying out Student’s t-test, while those among multiple groups by one-way ANOVA with post-hoc Dunnett’s multiple comparison. P < 0.05 stood for statistical significance.
Results
MiR-381-3p is downregulated within bone marrow in children with AML
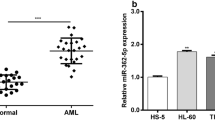
Zhang et al., as pointed out in the original review, have already demonstrated the expression of miR-381-3p in pediatric AML patients (Zhang et al. 2020). We examined miR-381-3p level within bone marrow in children with AML, as well as children receiving orthopedic surgery for correcting skeletal deformity (as controls). The qRT-PCR assay demonstrated that miR-381-3p level within the marrow of children with AML notably decreased compared with control group (Fig. 1A). Besides, its expression was significantly correlated with cytogenetics (Table 1). Moreover, miR-381-3p expression was also downregulated in AML cells (HL-60 and U937) compared to normal HS-5 cells (Fig. 1B).
miR-381-3p shows downregulation within in children with AML. RT-qPCR measured level of miR-381-3p in A bone marrow samples of AML patients (AML; n = 34) and normal participators (n = 21), and B AML cell lines including HL-60 and U937, as well as normal bone marrow stromal cell line HS-5. Results are displayed in a form of means ± SD from 3 separate assays. *P < 0.05; **P < 0.01 relative to children receiving orthopedic surgery for correcting skeletal deformity
miR-381-3p overexpression suppresses cell growth, induces cell cycle arrest, and promotes cell apoptosis
For exploring miR-381-3p’s effect on pediatric AML, we evaluated cell proliferation, cell cycle and apoptosis in AML cell lines HL-60 and U937 in response to the transfection of miR-381-3p mimic or inhibitor. Based on MTT assays, miR-381-3p upregulation markedly inhibited AML cell growth (Fig. 2A). However, miR-381-3p knockdown obviously promoted cell proliferation at the indicated time points in HL-60 and U937 cells (Fig. S1A). Afterwards, we conducted flow cytometry (FCM) for detecting cell apoptosis and cell cycle. Cells with miR-381-3p overexpression exhibited dramatically elevated G1-phase cell percentage, but significantly decreased S-phase cell percentage, which indicated that miR-381-3p overexpression induced cell cycle arrest at G1 phase (Fig. 2B), which was reversed by miR-381-3p silencing (Fig. S1B). Furthermore, after miR-381-3p mimics transfection, the cell apoptosis rate dramatically increased relative to NC (Fig. 2C), while the apoptosis was significantly inhibited following miR-381-3p knockdown (Fig. S1C).
miR-381-3p overexpression suppresses cell growth, and causes cell apoptosis and cell cycle arrest. miR-381-3p mimics or NC was transfected into HL-60 and U937 cells. A MTT assay was conducted to test cell viability. B FCM was conducted to analyze cell cycle distribution. C Cell apoptosis was measured by FCM. Results were expressed as means ± SD from 3 separate assays. *P < 0.05, **P < 0.01 compared with NC group
Overexpression of miR-381-3p represses cell invasion and migration
For investigating the role of miR-381-3p in AML cell motility, we analyzed HL-60 and U937 cell migration and invasion upon miR-381-3p overexpression or knockdown. As revealed by results of Transwell assay, miR-381-3p upregulation dramatically reduced HL-60 and U937 cell migration and invasion compared with NC group (Fig. 3A and B). On the contrary, the inhibition of miR-381-3p led to an increase in cell migration and invasion of HL-60 and U937 cells (Fig. S2).
ROCK1 serves as miR-381-3p’s target
For illustrating the role of miR-381-3p in repressing cell proliferation, migration, and invasion, its potential targets were identified by the TargetScan online software. We predicted ROCK1, frequently reported to be involved in various human malignancies, to be miR-381-3p’s target (Fig. 4A). For validating whether ROCK1 was modulated by directly binding miR-381-3p to the corresponding 3′ UTR, we cloned the ROCK1 3′ UTR fragment that contained miR-381-3p target sequences, or the relevant mutant fragment in firefly luciferase vectors. The results showed that miR-381-3p overexpression decreased while knockdown increased the luciferase activity in HL-60 and U937 cells transfected with the WT ROCK1 but not the activity of the MUT ROCK1 (Fig. 4B). According to Fig. 4C, ROCK1 expression decreased within miR-381-3p mimic-transfected cells relative to NC group.
ROCK1 serves as miR-381-3p’s direct target. A Prediction of miR-381-3p’s candidate-binding site in ROCK1 3′ UTR using online TargetScan software. B miR-381-3p mimic, inhibitor, or negative controls and a luciferase plasmid carrying the WT or MUT ROCK1 3′ UTR reporter vectors were transfected into HL-60 and U937 cells. After 48 h of transfection, the transfected cells were subjected to quantification of luciferase activity using a dual-luciferase reporter assay system. C ROCK1 level detected through WB analysis within miR-381-3p mimics or NC-transfected HL-60 and U937 cells. **P < 0.01 relative to NC group
Restoring ROCK1 expression reverses miR-381-3p’s suppression on cell growth, invasion, and migration
As ROCK1 is tightly related to cancer cell proliferation, invasion, and migration, ROCK1 expression within clinical specimens was determined through qRT-PCR. As a result, ROCK1 was markedly upregulated in the bone marrow of children with AML compared with children receiving orthopedic surgery for correcting skeletal deformity (Fig. 5A). For determining whether miR-381-3p’s inhibition on cell proliferation, invasion and migration were mediated by the downregulation of ROCK1, we transfected miR-381-3p overexpression cells using recombinant lentivirus carrying ROCK1 gene (Fig. 5B). As demonstrated by the results shown in Fig. 5C-E, overexpression of ROCK1 reversed the inhibition of cell proliferation, migration, and invasion ability by miR-381-3p upregulation. Besides, ROCK inhibitor Y-27632 and miR-381-3p overexpression have similar results with respect to cell proliferation, migration, and invasion (Fig. S3).
Rescuing ROCK1 expression reverses miR-381-3p overexpression’s inhibition on cell growth, invasion and migration. A ROCK1 level within the bone marrow of children with AML identified by RT-PCR. B ROCK1 level analyzed through WB assay within miR-381-3p mimics or NC-transfected HL-60 and U937 cells. C MTT assay conducted to detect cell proliferation. D–E Cell migration and invasion examined by Transwell assays. Results were displayed as means ± SD from 3 separate assays. **P < 0.01
Discussion
Recent researches have reported that miRNAs play a crucial role in pediatric AML. For example, as suggested by Ramamurthy et al., miR-155 level showed close relation to clinical outcome in pediatric AML (Ramamurthy et al. 2016). According to Emmrich et al., miR-9 was the tumor suppressor, which inhibited AML cell proliferation (Emmrich et al. 2014). Moreover, Zhang et al. discovered miR-125a downregulation within pediatric AML, while miR-125a inhibited the proliferation of HL-60 cells (Zhang et al. 2018). Emerging evidence revealed miR-381-3p usually inhibits the progression of diverse cancers, such as hepatocellular cancer (HCC) (Zhang et al. 2016), GC (Zhang et al. 2017), OSCC (Yang et al. 2017), colorectal cancer (CRC) (He et al. 2016), breast cancer (BC) (Xue et al. 2017), renal cancer (Chen et al. 2013), lung adenocarcinoma (LUAD) (Rothschild et al. 2012), ovarian cancer (Xia et al. 2016), and esophageal squamous cell carcinoma (ESCC) (Zhou et al. 2015). Recently, miR-381 has been shown to be downregulated in the bone marrow tissues of AML patients and the overexpression of miR-381 might inhibit the proliferation of AML cells by targeting HMGB1 (Zhang et al. 2020). However, the underlying mechanisms of it remain unexplored. In this study, we confirmed that the expression of miR-381-3p was significantly downregulated in AML samples and cell lines. Clinically, high miR-381-3p level was associated with cytogenetics in AML, which had also been previously demonstrated (Zhang et al. 2020). Functional studies concluded that miR-381-3p overexpression suppressed cell growth, inhibited cell invasion and migration, and promoted apoptosis and cell cycle arrest of AML cells, while miR-381-3p knockdown led to an opposite effect, indicating the feasibility of miR-381-3p to be the tumor suppressor of pediatric AML.
For further exploring miR-381-3p’s mechanism as the tumor suppressor of pediatric AML, we predicted its candidate targets using TargetScan online software. Through dual-luciferase reporter assay, we identified ROCK1, a tumor-associated gene, to be miR-381-3p’s direct target. ROCK1, an essential effector kinase in the downstream Rho GTPase, has an indispensable role in cytoskeleton reorganization and cell mobility (Cai et al. 2015; Akagi et al. 2014). ROCK1 has been increasingly suggested as an oncogene of diverse human tumors, including GC (Shin et al. 2014), non-small cell lung cancer (NSCLC) (Li et al. 2013; Vigil et al. 2012), bladder cancer (Wu et al. 2016; Majid et al. 2012), rectal cancer (Cai et al. 2015), and vulvar cancer (Akagi et al. 2014). Besides, ROCK1 is reported to be over-expressed in AML, and serves as the therapeutic target in AML (Wermke et al. 2015).
The current work observed that ROCK1 expression markedly increased within bone marrow in AML children. For better determining the association of miR-381-3p with ROCK1 expression, HL-60 and U937 cells overexpressing miR-381-3p were transfected with lentivirus carrying ROCK1 gene. According to our results, ROCK1 level was markedly decreased within HL-60 and U937 cells over-expressing miR-381-3p. Additionally, rescuing ROCK1 level reversed miR-381-3p’s inhibition on cell growth, invasion and migration. Furthermore, the suppression of ROCK1 expression using ROCK inhibitor was sufficient to reduce cell proliferation, migration, and invasion ability of AML cell lines. Based on our results, miR-381 was the tumor suppressor through directly targeting ROCK1. Some limitations of this study are noted. First, the clinical significance of miR-381-3p and ROCK1 has not been investigated in a larger cohort. Further 5-year follow-up for AML patients was needed. Second, further in vivo experiments were necessary for the confirmation of the function of miR-381-3p in AML cells. Third, further in-depth exploration was essential for potential signaling pathway through which miR-381-3p exhibited its tumor-related effects.
To sum up, this work demonstrates downregulation of miR-381-3p within pediatric AML, which targets ROCK1 and serves as the tumor suppressor. This work sheds novel lights on those molecular mechanisms in pediatric AML occurrence. Therefore, miR-381-3p is the possibly novel anti-AML therapeutic target in children.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Akagi EM, Lavorato-Rocha AM, Maia Bde M, Rodrigues IS, Carvalho KC, Stiepcich MM et al (2014) ROCK1 as a novel prognostic marker in vulvar cancer. BMC Cancer 14:822
Alcalay M, Tiacci E, Bergomas R, Bigerna B, Venturini E, Minardi SP et al (2005) Acute myeloid leukemia bearing cytoplasmic nucleophosmin (NPMc+ AML) shows a distinct gene expression profile characterized by up-regulation of genes involved in stem-cell maintenance. Blood 106(3):899–902
Bachas C, Schuurhuis GJ, Hollink IH, Kwidama ZJ, Goemans BF, Zwaan CM et al (2010) High-frequency type I/II mutational shifts between diagnosis and relapse are associated with outcome in pediatric AML: implications for personalized medicine. Blood 116(15):2752–2758
Brown P, Meshinchi S, Levis M, Alonzo TA, Gerbing R, Lange B et al (2004) Pediatric AML primary samples with FLT3/ITD mutations are preferentially killed by FLT3 inhibition. Blood 104(6):1841–1849
Cai SD, Chen JS, Xi ZW, Zhang LJ, Niu ML, Gao ZY (2015) MicroRNA144 inhibits migration and proliferation in rectal cancer by downregulating ROCK1. Mol Med Rep 12(5):7396–7402
Chen B, Duan L, Yin G, Tan J, Jiang X (2013) Simultaneously expressed miR-424 and miR-381 synergistically suppress the proliferation and survival of renal cancer cells–-Cdc2 activity is up-regulated by targeting WEE1. Clinics (Sao Paulo) 68(6):825–833
Djamai H, Berrou J, Dupont M, Kaci A, Ehlert JE, Weber H et al (2021) Synergy of FLT3 inhibitors and the small molecule inhibitor of LIM kinase1/2 CEL_amide in FLT3-ITD mutated acute myeloblastic leukemia (AML) cells. Leuk Res 100:106490
Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F et al (2014) miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia 28(5):1022–1032
He X, Wei Y, Wang Y, Liu L, Wang W, Li N (2016) MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1. Onco Targets Ther 9:1231–1239
Hu CB, Li QL, Hu JF, Zhang Q, Xie JP, Deng L (2014) miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac J Cancer Prev: APJCP 15(16):6543–6546
Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T (2002) Expression of ROCK-1 in human pancreatic cancer: its down-regulation by morpholino oligo antisense can reduce the migration of pancreatic cancer cells in vitro. Pancreas 24(3):251–257
Kroiss A, Vincent S, Decaussin-Petrucci M, Meugnier E, Viallet J, Ruffion A et al (2015) Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 34(22):2846–2855
Li J, Song Y, Wang Y, Luo J, Yu W (2013) MicroRNA-148a suppresses epithelial-to-mesenchymal transition by targeting ROCK1 in non-small cell lung cancer cells. Mol Cell Biochem 380(1–2):277–282
Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS et al (2012) MicroRNA-1280 inhibits invasion and metastasis by targeting ROCK1 in bladder cancer. PLoS ONE 7(10):e46743
Oke A, Pearce D, Wilkinson RW, Crafter C, Odedra R, Cavenagh J et al (2009) AZD1152 rapidly and negatively affects the growth and survival of human acute myeloid leukemia cells in vitro and in vivo. Can Res 69(10):4150–4158
Pession A, Masetti R, Rizzari C, Putti MC, Casale F, Fagioli F et al (2013) Results of the AIEOP AML 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood 122(2):170–178
Ramamurthy R, Hughes M, Morris V, Bolouri H, Gerbing RB, Wang YC et al (2016) miR-155 expression and correlation with clinical outcome in pediatric AML: a report from Children’s Oncology Group. Pediatr Blood Cancer 63(12):2096–2103
Raviraj V, Fok S, Zhao J, Chien HY, Lyons JG, Thompson EW et al (2012) Regulation of ROCK1 via Notch1 during breast cancer cell migration into dense matrices. BMC Cell Biol 13:12
Rothschild SI, Tschan MP, Jaggi R, Fey MF, Gugger M, Gautschi O (2012) MicroRNA-381 represses ID1 and is deregulated in lung adenocarcinoma. J Thorac Oncol 7(7):1069–1077
Rubnitz JE, Lacayo NJ, Inaba H, Heym K, Ribeiro RC, Taub J et al (2019) Clofarabine can replace anthracyclines and etoposide in remission induction therapy for childhood acute myeloid leukemia: the AML08 multicenter, randomized phase III trial. J Clin Oncol 37(23):2072–2081
Shalini V, Pushpan CK, Sindhu G, Jayalekshmy A, Helen A (2016) Tricin, flavonoid from Njavara reduces inflammatory responses in hPBMCs by modulating the p38MAPK and PI3K/Akt pathways and prevents inflammation associated endothelial dysfunction in HUVECs. Immunobiology 221(2):137–44
Shin JY, Kim YI, Cho SJ, Lee MK, Kook MC, Lee JH et al (2014) MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. PLoS ONE 9(1):e85205
Vigil D, Kim TY, Plachco A, Garton AJ, Castaldo L, Pachter JA et al (2012) ROCK1 and ROCK2 are required for non-small cell lung cancer anchorage-independent growth and invasion. Can Res 72(20):5338–5347
Wang W, Chen M, Gao Y, Song X, Zheng H, Zhang K et al (2018) P2Y6 regulates cytoskeleton reorganization and cell migration of C2C12 myoblasts via ROCK pathway. J Cell Biochem 119(2):1889–1898
Wang Y, Zhao W, Fu Q (2013) miR-335 suppresses migration and invasion by targeting ROCK1 in osteosarcoma cells. Mol Cell Biochem 384(1–2):105–111
Wang Z, Sun L, Liang S, Liu ZC, Zhao ZY, Yang J et al (2019) GPER stabilizes F-actin cytoskeleton and activates TAZ via PLCbeta-PKC and Rho/ROCK-LIMK-Cofilin pathway. Biochem Biophys Res Commun 516(3):976–982
Wermke M, Camgoz A, Paszkowski-Rogacz M, Thieme S, von Bonin M, Dahl A et al (2015) RNAi profiling of primary human AML cells identifies ROCK1 as a therapeutic target and nominates fasudil as an antileukemic drug. Blood 125(24):3760–3768
Wu D, Niu X, Pan H, Zhou Y, Qu P, Zhou J (2016) MicroRNA-335 is downregulated in bladder cancer and inhibits cell growth, migration and invasion via targeting ROCK1. Mol Med Rep 13(5):4379–4385
Xia B, Li H, Yang S, Liu T, Lou G (2016) MiR-381 inhibits epithelial ovarian cancer malignancy via YY1 suppression. Tumour Biol: J Int Soc Oncodevelopmental Biol Med 37(7):9157–9167
Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P (2017) miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother = Biomed Pharmacother 86:426–33
Yang X, Ruan H, Hu X, Cao A, Song L (2017) miR-381-3p suppresses the proliferation of oral squamous cell carcinoma cells by directly targeting FGFR2. Am J Cancer Res 7(4):913–922
Zampini M, Tregnago C, Bisio V, Simula L, Borella G, Manara E et al (2018) Epigenetic heterogeneity affects the risk of relapse in children with t(8;21)RUNX1-RUNX1T1-rearranged AML. Leukemia 32(5):1124–1134
Zhang M, Huang S, Long D (2017) MiR-381 inhibits migration and invasion in human gastric carcinoma through downregulatedting SOX4. Oncol Lett 14(3):3760–3766
Zhang P, Sun D, Sun X, Li H (2020) Clinical significance of dysregulation of miR-381 in pediatric acute myeloid leukemia. Eur J Med Res 25(1):42
Zhang Q, Zhao S, Pang X, Chi B (2016) MicroRNA-381 suppresses cell growth and invasion by targeting the liver receptor homolog-1 in hepatocellular carcinoma. Oncol Rep 35(3):1831–1840
Zhang Y, Liu Y, Xu X (2018) Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J Cell Biochem 119(7):6296–6308
Zheng B, Liang L, Wang C, Huang S, Cao X, Zha R et al (2011) MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res: Off J Am Assoc Cancer Res 17(24):7574–7583
Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J et al (2015) MicroRNA-381 increases radiosensitivity in esophageal squamous cell carcinoma. Am J Cancer Res 5(1):267–277
Zwaan CM, Kolb EA, Reinhardt D, Abrahamsson J, Adachi S, Aplenc R et al (2015) Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol 33(27):2949–2962
Funding
This study was funded by grants from Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2020KY254) and Ningbo Natural Science Foundation (2018A05).
Author information
Authors and Affiliations
Contributions
QY and QY were responsible for designing and performing experiments. QD and CL were in charge of analyzing data. GX played a role in enrolling cases and measuring RNA expression within clinical samples. QY was in charge of study initiation and manuscript writing. The authors approved the eventual manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent for participation
The Ethics Committee of Ningbo First Hospital of Zhejiang University approved human tissue utilization. All patients provided informed consents.
Conflict of interest
The authors declare no competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

ESM 1
Figure S1. miR-381-3p knockdown promotes cell growth, and inhibits apoptosis and cell cycle arrest. miR-381-3p inhibitor or inhibitor NC was transfected into HL-60 and U937 cells. (A) MTT assay was conducted to test cell viability. (B) FCM was conducted to analyze cell cycle distribution. (C) Cell apoptosis was measured by FCM. Results were expressed as means ± SD from 3 separate assays. *P < 0.05, **P < 0.01 compared with inhibitor NC group. (PNG 421 kb)

ESM 2
Figure S2. miR-381-3p knockdown promotes cell migration and invasion. miR-381-3p inhibitor or inhibitor NC were transfected into HL-60 and U937 cells. Transwell assays were conducted to assess (A) cell migration and (B) invasion. Results are displayed as means ± SD from 3 separate assays. **P < 0.01 relative to inhibitor NC group. (PNG 2015 kb)

ESM 3
Figure S3. ROCK inhibitor Y-27632 inhibits AML cell migration and invasion. miR-381-3p mimics or ROCK inhibitor Y-27632 were transfected into HL-60 and U937 cells, respectively. (A) MTT assay was conducted to test cell viability. Transwell assays were conducted to assess cell migration (B) and invasion (C). Results are displayed as means ± SD from 3 separate assays. **P < 0.01 relative to NC group. (PNG 1368 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, Q., Ying, Q., Dai, Q. et al. Tumor-suppressing effects of miR-381-3p in pediatric acute myeloid leukemia via ROCK1 downregulation. Funct Integr Genomics 23, 43 (2023). https://doi.org/10.1007/s10142-022-00950-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-022-00950-9